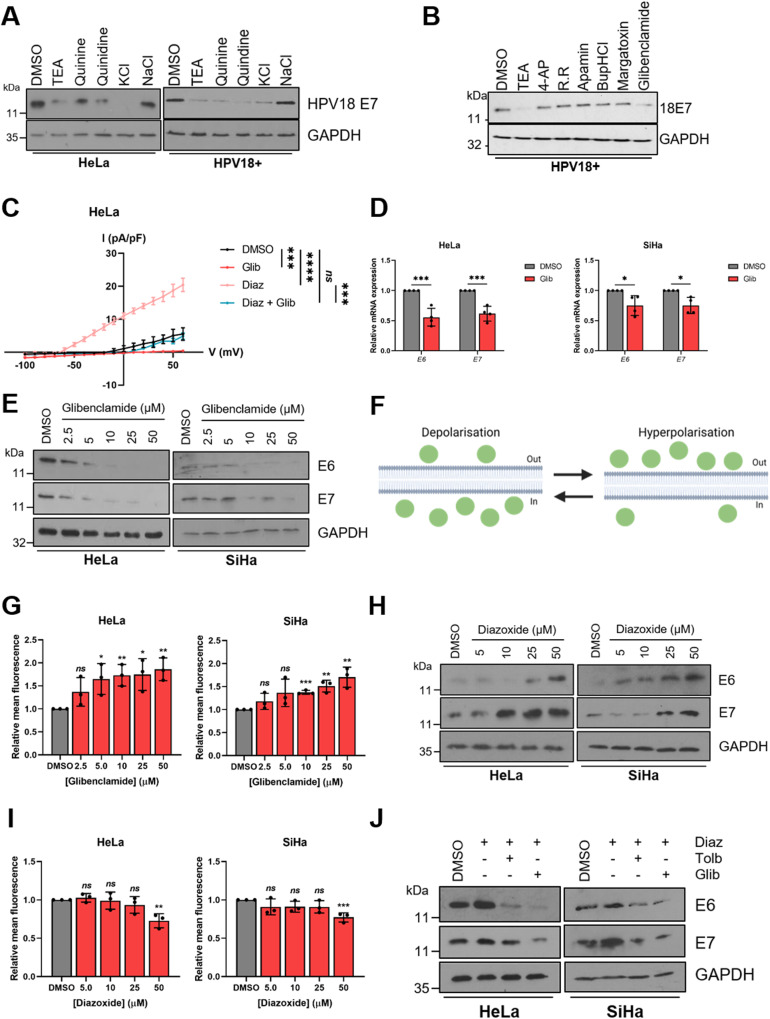
Fig. 1. KATP channels are important for HPV gene expression.
A Representative western blots for E7 expression in HeLa cells and primary human keratinocytes containing HPV18 episomes treated with DMSO, a broadly-acting K+ channel inhibitor (25 mM tetraethylammonium (TEA), 100 µM quinine, 100 µM quinidine or 70 mM KCl) or 70 mM NaCl. GAPDH served as a loading control. B Representative western blots for E7 expression in HPV18+ primary keratinocytes treated with DMSO, 25 mM TEA or one of a panel of class-specific K+ channel inhibitors (2 mM 4-aminopyridine (4-AP), 50 µM ruthenium red (RR), 10 nM apamin, 20 µM bupivacaine hydrochloride (BupHCl), 10 nM margatoxin or 50 μM glibenclamide). GAPDH served as a loading control. C Mean current density-voltage relationships for K+ currents in HeLa cells treated with DMSO, diazoxide (50 μM), glibenclamide (10 μM), or both diazoxide and glibenclamide (n = 5 for all treatments). D Expression levels of E6 and E7 mRNA in HeLa and SiHa cells treated with glibenclamide (10 μM) measured by RT-qPCR (n = 4 for all treatments). Samples were normalised against U6 mRNA levels. E Representative western blots of E6 and E7 expression in HeLa and SiHa cells treated with increasing doses of glibenclamide. GAPDH served as a loading control. F Schematic illustrating the plasma membrane permeability of DiBAC4(3). Figure created using BioRENDER.com. G Mean DiBAC4(3) fluorescence levels in HeLa and SiHa cells treated with increasing dose of glibenclamide. Samples were normalised to DMSO controls. H Representative western blots of E6 and E7 expression in HeLa and SiHa cells serum starved for 24 h (to reduce basal E6/E7 expression) prior to treatment with increasing doses of diazoxide. GAPDH served as a loading control. I Mean DiBAC4(3) fluorescence levels in HeLa and SiHa cells treated with increasing dose of diazoxide. Samples were normalised to DMSO control. J Representative western blots of E6 and E7 expression in HeLa and SiHa cells treated with diazoxide (50 μM) alone or in combination with glibenclamide (10 μM) or tolbutamide (200 μM). Bars represent means ± standard deviation (SD) of three biological replicates (unless stated otherwise) with individual data points displayed where possible. Ns not significant, *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t test).