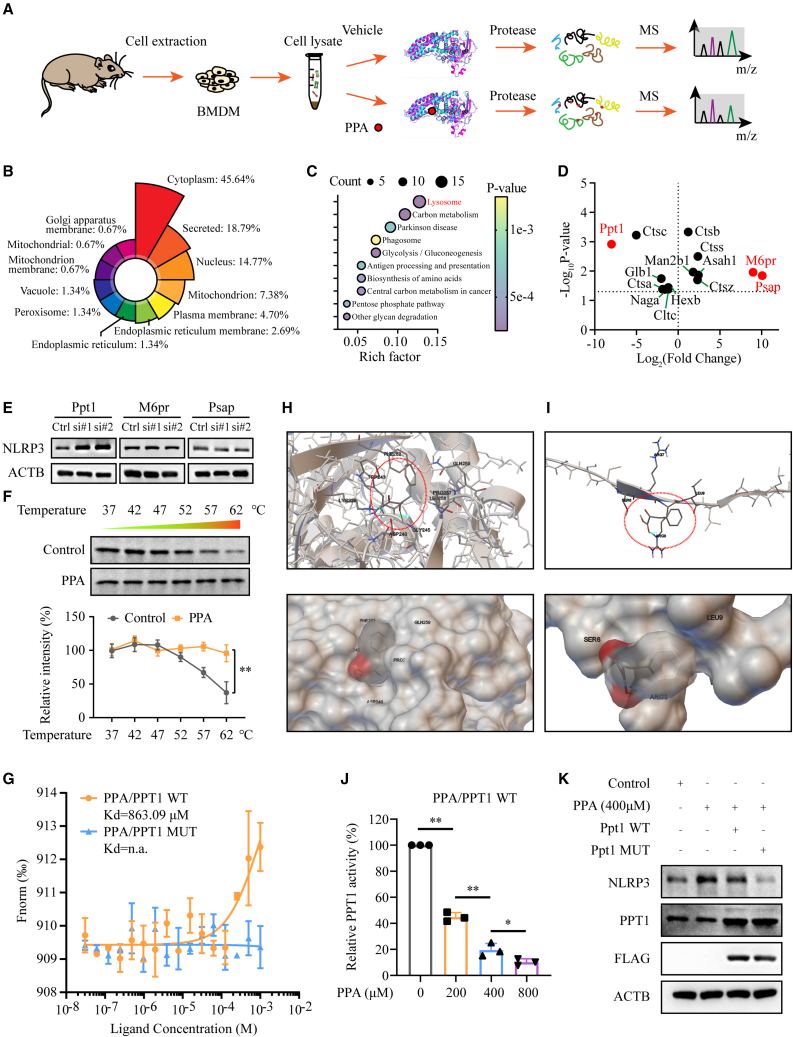
Figure 4.
Identification of potential phenylpyruvate targets in macrophages using LiP-SMap
(A) Flow chart depicting the LiP-SMap assay. The LiP-SMap assay is a chemical proteomics screening method for investigating potential protein-metabolite interactions.
(B) The rose chart reflects the proportion of differentially expressed polypeptide-dependent proteins mapped in each subcellular location.
(C) The bubble plot shows the KEGG pathway enrichment analysis. The abscissa represents the enrichment value, and the ordinate represents the enriched KEGG pathways. The larger the circle is, the more the differentially expressed polypeptide-dependent proteins in the mapping pathway are enriched. The color of the circle indicates the p value.
(D) A spot chart showing the differential proteins mapped in the lysosome pathway. Fold changes greater than 2 or less than 0.5 both indicate potential interactions between proteins and metabolites.
(E) Immunoblot analysis of NLRP3 expression levels in BMDMs with knockdown of Ppt1, M6pr, and Psap (n = 3).
(F) Cellular thermal shift assay and statistical analysis of PPT1 in BMDMs treated with or without phenylpyruvate (n = 3).
(G) Microscale thermophoresis assay of phenylpyruvate binding to the PPT1 WT or MUT protein.
(H) Representative images of autodocking for phenylpyruvate to the principal chain structure of the PPT1 protein.
(I) Representative images of autodocking for phenylpyruvate to the signal domain of the PPT1 protein.
(J) Enzymatic activity measurement of PPT1 incubated with increasing phenylpyruvate concentrations (n = 3).
(K) BMDMs were treated with phenylpyruvate and then transfected with the Ppt1 WT plasmid or the MUT plasmid for 24 h. Immunoblot analysis showing NLRP3 expression in BMDMs (n = 3). Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01; n.a., not applicable.