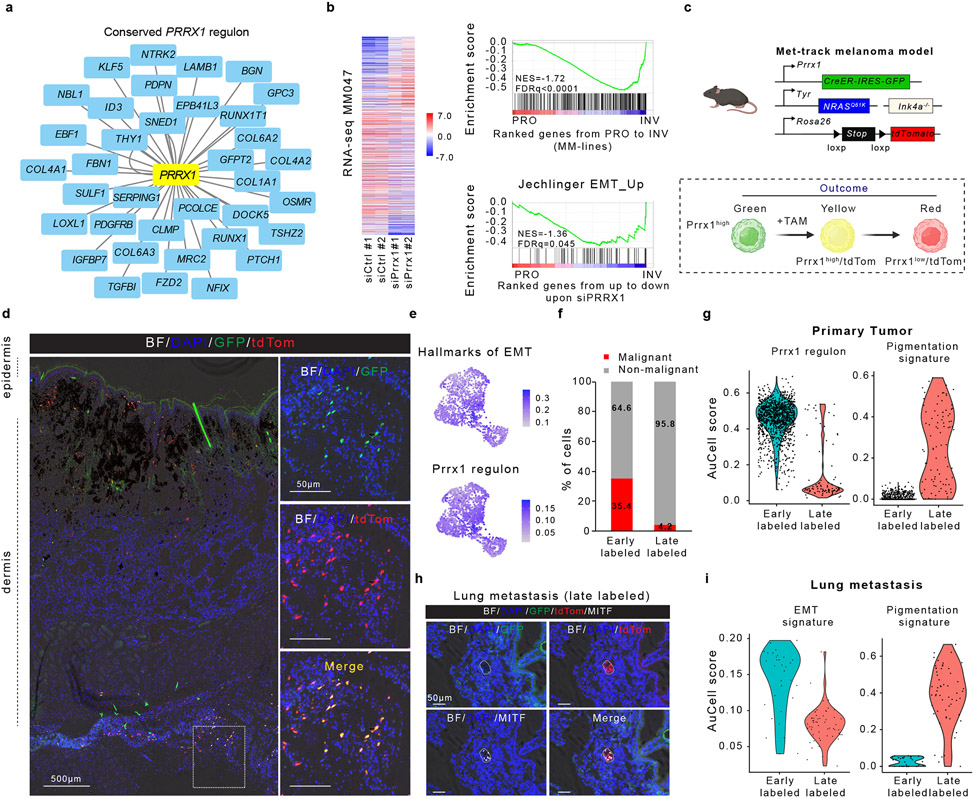
Figure 4: Temporal single-cell tracing identifies a population of melanoma cells that fuels metastasis but not primary tumour growth.
a, Conserved (mouse and human) PRRX1 regulon (see Methods). b, Left panel, heatmap of differentially expressed genes upon silencing of PRRX1 in MM047 cells (bulk RNAseq, n=2 biological replicates). Right panel, GSEA shows downregulation of mesenchymal-like signature upon siPRRX1 (MM-lines and Jechlinger_EMT_UP gene sets). c, Schematic representation of the Met-Track mouse model and possible lineage tracing outcomes following Tamoxifen (TAM) exposure. Schematic made with Biorender.com. d, Confocal images of a primary tumour 4 weeks post-TAM. Insets showing the majority of GFP/tdTomato-double positive cells in the deepest dermal part of the lesion. BF, Bright Field (n=4 independent mice). e UMAP shows hallmarks of EMT and PRRX1 regulon activity (AUCell score) in tdTomato+ cells 2 days (early labelled) post-TAM. f, Ratio malignant versus non-malignant (as determined by scRNA-seq) of FACS-sorted tdTomato+ fraction from primary tumours 2 days (early labelled) and 30-days (late labelled) post-TAM. g, Expression of PRRX1 regulon and pigmentation signatures (determined by scRNA-seq) of FACS-sorted tdTomato+ fraction 2 days (early labelled) and 30-days (late labelled) upon last TAM administration. h, Confocal images of a lung micro-metastasis 4 weeks post-TAM, immunostained for MITF (white). Red signal is from tdTomato expression; GFP (green) signal. BF, Bright Field (n=3 independent mice). i, Expression of PRRX1 regulon and pigmentation gene signatures (as determined by scRNA-seq) of FACS-sorted tdTomato+ fraction from lung metastatic lesions 2 days (early labelled) and 30-days (late labelled) post-TAM.