Fig. 1 |. A model for BFCN fate specification.
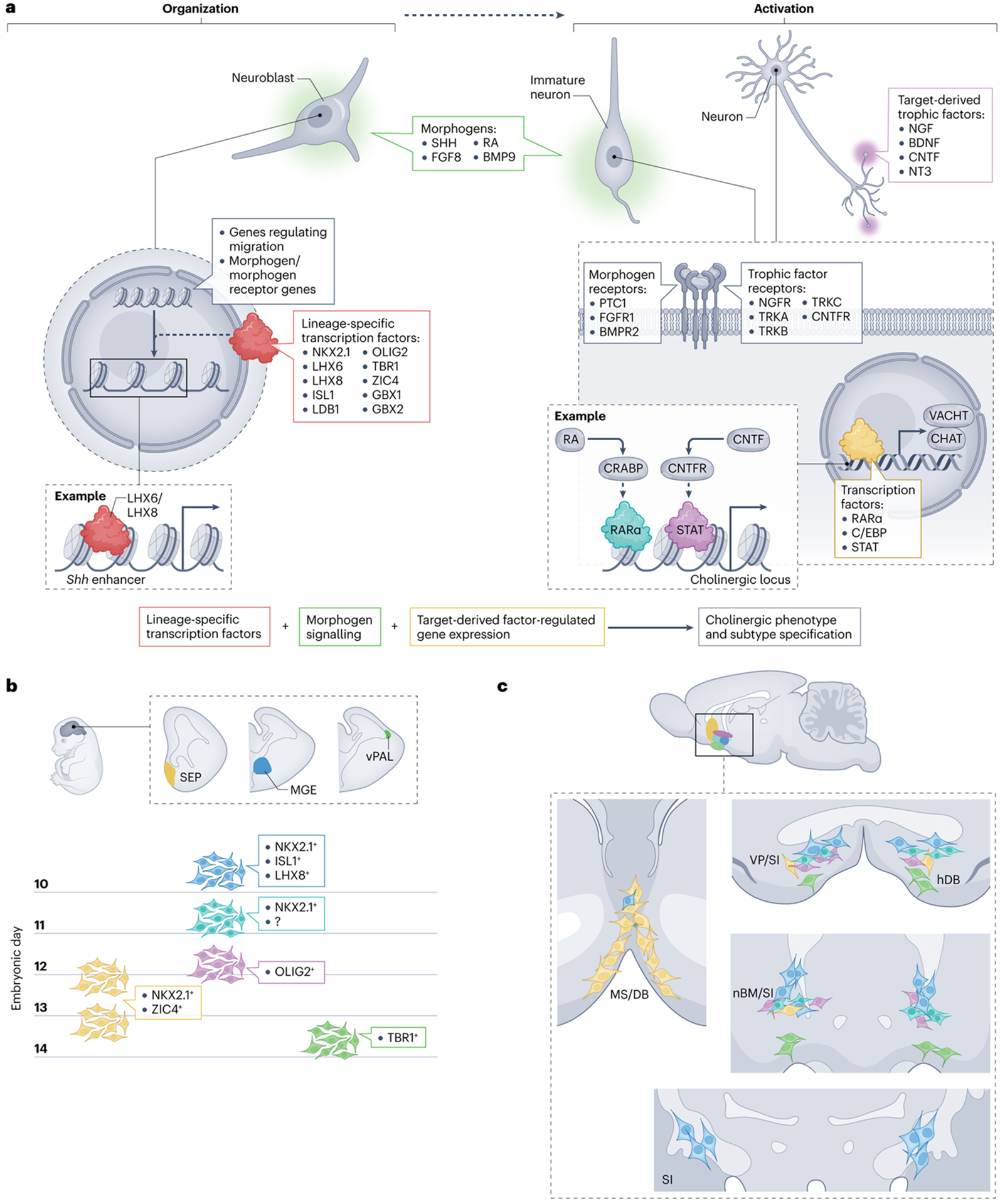
A proposed model for cholinergic neuron fate commitment based on evidence from genetic studies (Table 1), common developmental mechanisms among cholinergic cell populations and trends in developmental regulation43–111. a, The model proposes that lineage-specific transcription factors ‘organize’ the genome to regulate the expression of genes involved in the migration (radial or tangential) of immature neurons and/or neuroblasts and the expression of receptors for morphogens and trophic factors. Exposure to morphogens present in the basal forebrain or (later in development) to trophic factors present in the terminal fields ‘activates’ the genome. For example, the cholinergic gene locus has been shown to contain cis-regulatory elements that respond to retinoic acid (RA) signalling and a ciliary neurotrophic factor (CNTF) response element, STAT, that responds to CNTF signalling, meaning that treatment with either RA or CNTF increases choline acetyltransferase (CHAT) and vesicular acetylcholine transporter (VACHT) expression. b, The different germinal zones in the embryonic brain that give rise to basal forebrain cholinergic neurons (BFCNs) and our current understanding of the timing of the birth of the neurons in each of these zones and some of the contributing regulators. These regulators shown were selected on the basis of their expression in the progenitor zones and may represent master regulators. We propose that the earliest-born BFCNs (shown in blue) are generated from progenitors in the medial ganglionic eminence (MGE) and rely on the transcription factors NKX2.1, LHX8 and ISL1 for their specification, whereas subsequent generations of neurons born in this domain (shown in turquoise and purple) do not rely strongly on LHX8. The latest population to be derived from this zone is proposed to require OLIG2; however, it is also possible that this population is derived from a neighbouring germinal zone such as the anterior entopeduncular area or preoptic area. Most late-born BFCNs (shown in yellow) are generated in the septum (SEP) from NKX2.1-expressing and ZIC4-expressing progenitors and rely on NKX2.1 expression for their specification. However, one late-born population that expresses TBR1 (shown in green) migrates into the basal forebrain from the ventral pallium (vPAL). c, The proposed final location of the BFCNs derived from the progenitor populations proposed in part b. We propose that early-born MGE-derived progenitors produce the large cholinergic neurons that reside in the substantia innominata (SI), nucleus basalis of Meynert (nBM) and ventral pallidum (VP) (blue), whereas subsequently generated progenitors might produce the smaller cholinergic neurons in these regions (turquoise). The OLIG2-expressing lineage might generate the more medially located cholinergic neurons in these regions (purple). Late-born septal progenitors differentiate into large neurons that continue to populate the medial septum (MS) and diagonal band (DB) and migrate tangentially to populate the basal forebrain and striatum (yellow). Finally, the TBR1-expressing population (green) that arrives from the vPAL represents the most ventrally located neurons within the horizontal diagonal band (hDB), SI, nBM and VP. CNTFR, ciliary neurotrophic factor receptor; NGF, nerve growth factor; NT3, neurotrophin 3; SHH, sonic hedgehog protein.
