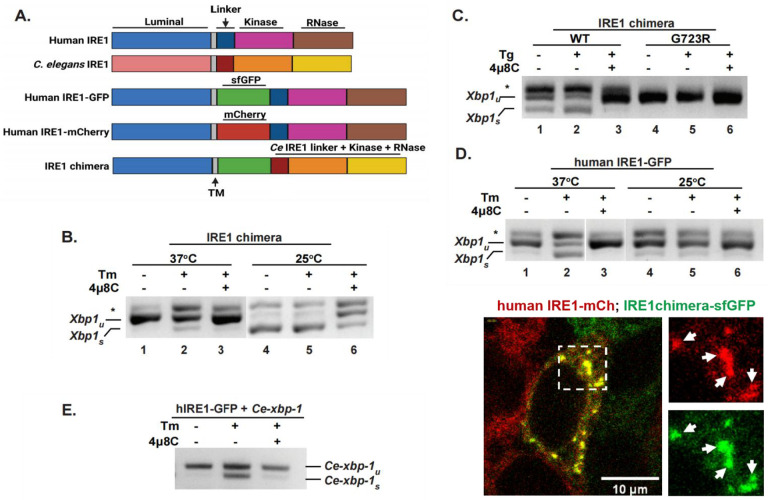
Figure 1. C. elegans cytosolic domains recognize human targets and are catalytically active in human cells.
(A) Schemes of domain organization in the human and C. elegans IRE1 proteins, and the constructs used in this work. The IRE1 chimera is composed of human lumenal and transmembrane (TM) domains (amino acid 1 to 494 of human IRE1α), a super-folder GFP (sfGFP), and C. elegans IRE1 cytosolic portion including the linker region and the kinase/RNase domain (amino acid 456 to 967 of C. elegans IRE1). (B, C) IRE1 chimera is active and kinase-dependent. RT-PCR assay was used to measure Xbp1 splicing in HAP1 IRE1KO cells reconstituted with an IRE1 chimera, either wildtype or G723R kinase-dead mutant. Cells were treated with 4μg/ml tunicamycin (Tm) or 2.5 μM thapsigargin (Tg) for 4hrs, either at 25°C or 37°C, as indicated. IRE1 inhibitor 4μ8C was used at 16 μM. Xbp-1u: unspliced Xbp1; Xbp1s: spliced Xbp1; *, non-specific PCR product, likely a hybrid of spliced and unspliced products. (D) RNase activity of human IRE1 is also temperature-dependent. HAP1 IRE1KO cells stably transfected with human IRE1-GFP protein were treated as in B. Human IRE1 is nearly inactive at 25°C. (E) Human IRE1 recognizes and cleaves C. elegans xbp-1. HAP1 IRE1KO cells stably expressing human IRE1-GFP protein were transfected with C. elegans full-length xbp-1 and treated as in C. (F) IRE1 chimera is able to oligomerize and form clusters with human IRE1 under strong ER stress. HAP1 IREKO cells expressing both the human IRE1-mCherry and IRE1 chimera (containing sfGFP) were treated with SubAB (0.1μg/ml) for 4hrs. Cells were imaged at 25°C, confocal projection of a z-stack is shown for a representative cell containing clusters. Arrows: examples of clusters.