Abstract
Soil salinity severely hampers agricultural productivity. Under salt stress, excess Na+ accumulation causes cellular damage and plant growth retardation, and membrane Na+ transporters play central roles in Na+ uptake and exclusion to mitigate these adverse effects. In this study, we performed sos1 suppressor mutant (named sup) screening to uncover potential genetic interactors of SOS1 and additional salt tolerance mechanisms. Map-based cloning and sequencing identified a group of mutants harboring dominant gain-of-function mutations in the vacuolar Na+/H+ antiporter gene AtNHX1. The gain-of-function variants of AtNHX1 showed enhanced transporter activities in yeast cells and increased salt tolerance in Arabidopsis wild type plants. Ion content measurements indicated that at the cellular level, these gain-of-function mutations resulted in increased cellular Na+ accumulation likely due to enhanced vacuolar Na+ sequestration. However, the gain-of-function suppressor mutants showed reduced shoot Na+ but increased root Na+ accumulation under salt stress, indicating a role of AtNHX1 in limiting Na+ translocation from root to shoot. We also identified another group of sos1 suppressors with loss-of-function mutations in the Na+ transporter gene AtHKT1. Loss-of-function mutations in AtHKT1 and gain-of-function mutations in AtNHX1 additively suppressed sos1 salt sensitivity, which indicates that the three transporters, SOS1, AtNHX1 and AtHKT1 function independently but coordinately in controlling Na+ homeostasis and salt tolerance in Arabidopsis. Our findings provide valuable information about the target amino acids in NHX1 for gene editing to improve salt tolerance in crops.
Supplementary Information
The online version contains supplementary material available at 10.1007/s44154-021-00014-1.
Keywords: sos1 suppressor, AtHKT1, AtNHX1, Gain-of-function, Salt tolerance
Introduction
Crop productivity and plant health are severely compromised by high soil salinity. Salt stress leads to disequilibrium of sodium (Na+) and nutritional ions in plant cell, which could result in cell death and plant destruction. Specifically, excess Na+ can cause disruptions in normal enzymatic activity and impair cellular metabolism. Therefore, maintenance of ion homeostasis in the cell is required for better plant health under salt stress. Previous studies have identified genes that help Na+ homeostasis and promote cellular growth and survival (Blumwald 2000; Hasegawa et al. 2000; Zhu 2003; Assaha et al. 2017; Basu et al. 2021). Central to these are plasma membrane and vacuolar Na+ transporters that directly control cellular Na+ concentration and subcellular Na+ allocation.
SOS1 (SALT-OVERLY-SENSITIVE1) encodes a plasma membrane Na+/H+ antiporter, and its functional mutant sos1 is hypersensitive to NaCl (Shi et al. 2000). Conversely, overexpression of SOS1 increases salt tolerance by maintaining Na+ homeostasis under high salinity (Shi et al. 2003). SOS1 is phylogenetically related to other plasma membrane Na+/H+ antiporters such as Schizosaccharomyces pombe SOD2 and Saccharomyces cerevisiae NHA1 and mediates cellular Na+ efflux (Jia et al. 1992; Prior et al. 1996; Shi et al. 2000). At the whole plant level, SOS1 plays a critical role in controlling long-distance Na+ transport from root to shoot (Shi et al. 2002; El Mahi et al. 2019). Its activity is induced by salt stress signals that are transduced by the calcium (Ca2+)- dependent SOS signaling pathway. This pathway comprises of the Ca2+-binding protein SOS3 physically interacting with and activating the protein kinase SOS2, which, upon forming a protein kinase complex, phosphorylates and activates SOS1 by releasing its autoinhibition (Qiu et al. 2002; Zhu 2003; Quintero et al. 2011). Additionally, several other possible regulatory motifs are present in SOS1, which includes a binding motif for cyclic nucleotides and an interacting domain for the signaling protein RCD1 (Katiyar-Agarwal et al. 2006; Gong et al. 2020).
AtHKT1 is a plasma membrane transporter identified to mediate Na+ uptake (Rubio et al. 1995; Uozumi et al. 2000). Its importance in Na+ homeostasis was established by its genetic interaction with the SOS pathway genes. Loss-of-function hkt1 alleles suppress the salt hypersensitivity of sos1, sos2, and sos3 mutants (Rus et al. 2001). Genetic and physiological evidence supports that AtHKT1 controls Na+ translocation in plants, and it also plays a role in the regulation of plant K+ nutrition (Rus et al. 2004). It was initially proposed that AtHKT1 mitigates Na+ accumulation in shoots by recirculating Na+ to the roots via the phloem transport system (Maser et al. 2002; Berthomieu et al. 2003). However, AtHKT1 was later found to selectively unload Na+ from xylem vessels to xylem parenchyma cells, and thus reducing Na+ translocation to shoots and protecting photosynthetic leaves from salinity stress (Sunarpi et al. 2005). AtHKT1;1 was further demonstrated to independently control Na+ accumulation in root and Na+ retrieval from the xylem (Davenport et al. 2007). Specifically, overexpression of AtHKT1;1 in the mature root stele increased salt tolerance of the transgenic Arabidopsis by reducing Na+ accumulation in the shoot due to increased influx of Na+ into stellar root cells (Møller et al. 2009). The HKT1 system presents an evolutionally important salt sequestration mechanism in plants, as increasing number of homologs were found and characterized in other species, especially in agriculturally important monocots such as rice and wheat (Hamamoto et al. 2015; Ali et al. 2019; Gong et al. 2020). In fact, allelic variations in HKT1 and its homologs display differences in salinity tolerance within a species (Campbell et al. 2017; Busoms et al. 2018).
The vacuolar Na+/H+ antiporter gene AtNHX1 has been found to improve salt tolerance in Arabidopsis and tomato through overexpression (Apse et al. 1999; Zhang and Blumwald 2001). AtNHX1 is similar sequence-wise with the mammalian NHE and the yeast NHX1 Na+/H+ exchangers (Counillon and Pouyssegur 2000). AtNHX1 complements the yeast nhx1 mutant and suppresses its Na+/Li+ sensitive phenotype, indicating that AtNHX1 functions as an endosomal Na+/H+ antiporter (Gaxiola et al. 1999; Quintero et al. 2000). More importantly, NHX1 overexpression improved salinity tolerance in multiple plant species (Shi and Zhu 2002; Chen et al. 2007; Zhang and Shi 2013; Sahoo et al. 2016; Nguyen et al. 2019). As a vacuolar membrane Na+/H+ antiporter, AtNHX1 compartmentalizes Na+ to the vacuole and prevents toxic Na+ accumulation in the cytoplasm and aids in acclimation to salt stress (Blumwald 2000; Hasegawa et al. 2000; Gong et al. 2020). Vacuolar Na+ compartmentalization and the accumulation of organic solutes in the cytosol and other organelles play crucial roles in maintaining cellular water status and intracellular osmotic status (Blumwald et al. 2000). Na+ transport across the plasma membrane and tonoplast is dependent on the proton (H+) electrochemical potential established by H+-translocating pumps. Upregulation of H+-pyrophosphatase pump (H+-PPase) has been found to increase salt and drought tolerance in plants (Gaxiola et al. 2001; Park et al. 2005; Brini et al. 2007; Shen et al. 2015). In addition, the H+-pyrophosphatase AVP1 also plays a role in auxin transport and consequently auxin-dependent development (Li et al. 2005). Like AVP1, AtNHX1 has demonstrated other physiological functions such as control of cellular pH and development. For example, a null mutation in the homolog of AtNHX1 in morning glory (Ipomoea nil) disrupts the vacuolar pH of corolla cells, leading to altered flower color (Fukada-Tanaka et al. 2000; Yamaguchi et al. 2001). Other AtNHX1 homologs in Arabidopsis and other plant species are also reported to function in regulating pH, K+ homeostasis, vesicle trafficking, and cell expansion, all of which have substantial importance in plant development (Rodriguez-Rosales et al. 2009; Bassil et al. 2011; Reguera et al. 2015; Qiu 2016).
In this study, we present the identification and characterization of sos1 suppressors. Gain-of-function (dominant) mutations in AtNHX1 and loss-of-function (recessive) mutations in AtHKT1 that suppress the salt sensitivity of sos1–1 were identified. These suppressor mutations prevented cytosolic Na+ accumulation and mitigated Na+ translocation to the shoot and thus conferring enhanced salt tolerance in sos1–1. We further demonstrated that single gain-of-function amino acid substitutions in AtNHX1 provide improved salt tolerance in Arabidopsis, which paves a new avenue for crop improvement in salt tolerance by gene editing in the future. Our findings further highlight the complex interaction among the three crucial transporters for salt tolerance in Na+ homeostasis and salt tolerance in plants.
Results
Identification of sos1 suppressors
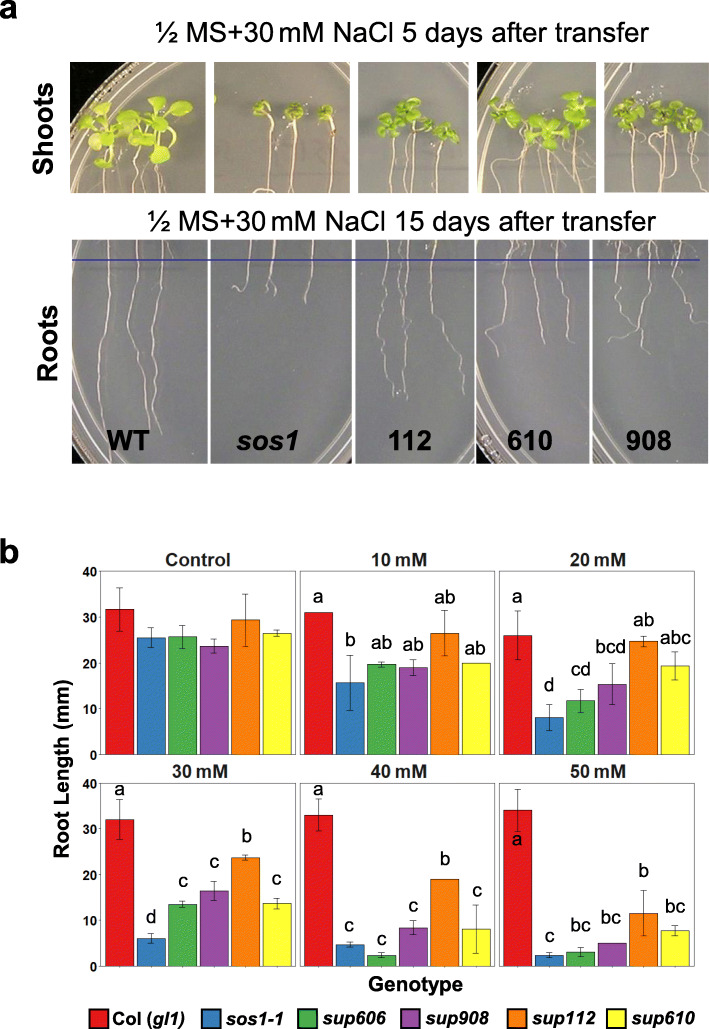
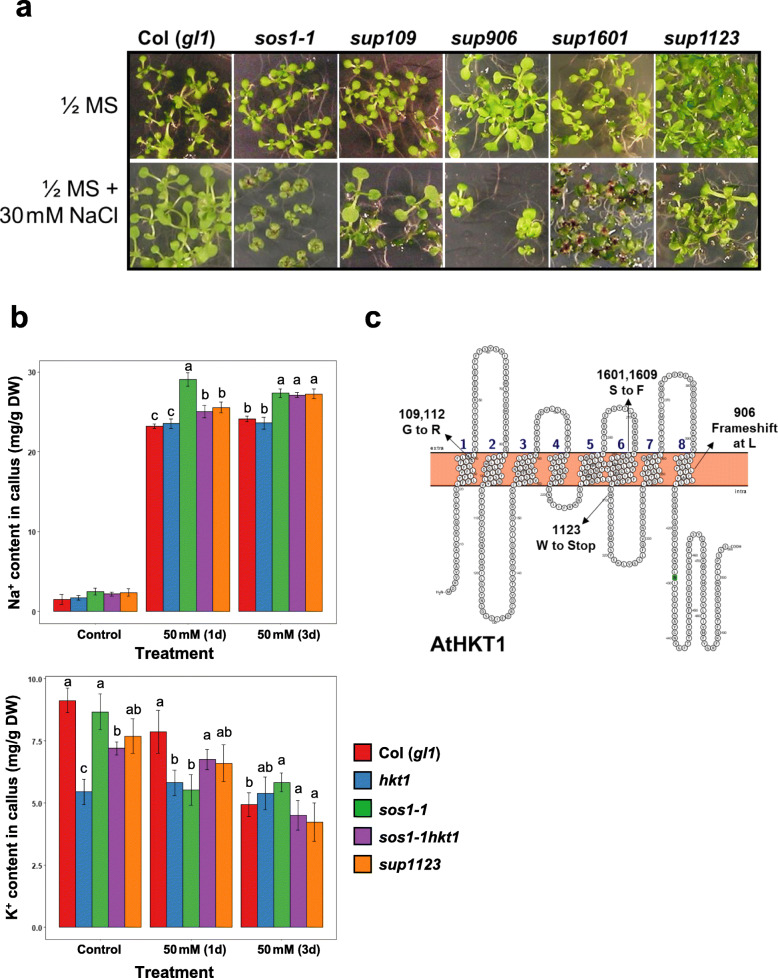
We performed a genetic screening for mutants suppressing the salt sensitivity of sos1 to identify genes coordinately working with the plasma membrane Na+/H+ antiporter SOS1 in conferring salinity tolerance. The sos1–1 allele carrying a 14 bp deletion (Shi et al. 2000) was used as the genetic background to avoid sos1 revertant mutants and recover additional mutations suppressing sos1–1. From a screening of ~ 250,000 M2 individuals of EMS-mutagenized sos1–1 and re-screening of putative sos1 suppressor lines, more than 40 lines were verified to be bona fide sos1 suppressors (designated as sup) and not sos1 revertant lines since no additional mutations were found in the sos1–1 mutant gene. Figure 1a shows the sos1 suppression phenotype of three selected sup mutants. The sup lines showed improved growth of both shoot and root when the growing media was supplemented with 30 mM NaCl. Root length measurements indicated that the four tested sup lines showed differences on the capacity of suppression of sos1–1 salt sensitivity (Fig. 1b). The sup112 suppressor mutant displayed strongest suppression of sos1–1 salt sensitivity in root growth, and the other three suppressors only showed clear suppression at 30 mM NaCl. However, sup112 exhibited weaker suppression in leaves than other two suppressors and sup610 showed better leaf growth among the tested suppressors (Fig. 1a). These results implied that multiple genes may be involved in suppressing the salt sensitivity of sos1–1, and these genes may have different tissue-specific modes of activity in response to salt stress.
Fig. 1.
Phenotypes of sos1 suppressor mutants. a Shoot and root growth of wild type (WT), sos1–1, and selected sos1 suppressor mutants. Five-day-old seedlings grown in ½ MS agar media (1.2% agar) were transferred to the same media supplemented with 30 mM NaCl. b Quantification of the root length of WT, sos1–1 and sos1 suppressor mutants grown in ½ MS agar media supplemented with different concentrations of NaCl. Bars represent means, and error bars represent standard errors (n = 5). Statistical significance among genotypes within treatments was computed with ANOVA and Tukey’s post-hoc HSD test (P < 0.05) and different statistical groups are represented by letters
Gain-of-function mutations in AtNHX1 suppress sos1 salt sensitivity
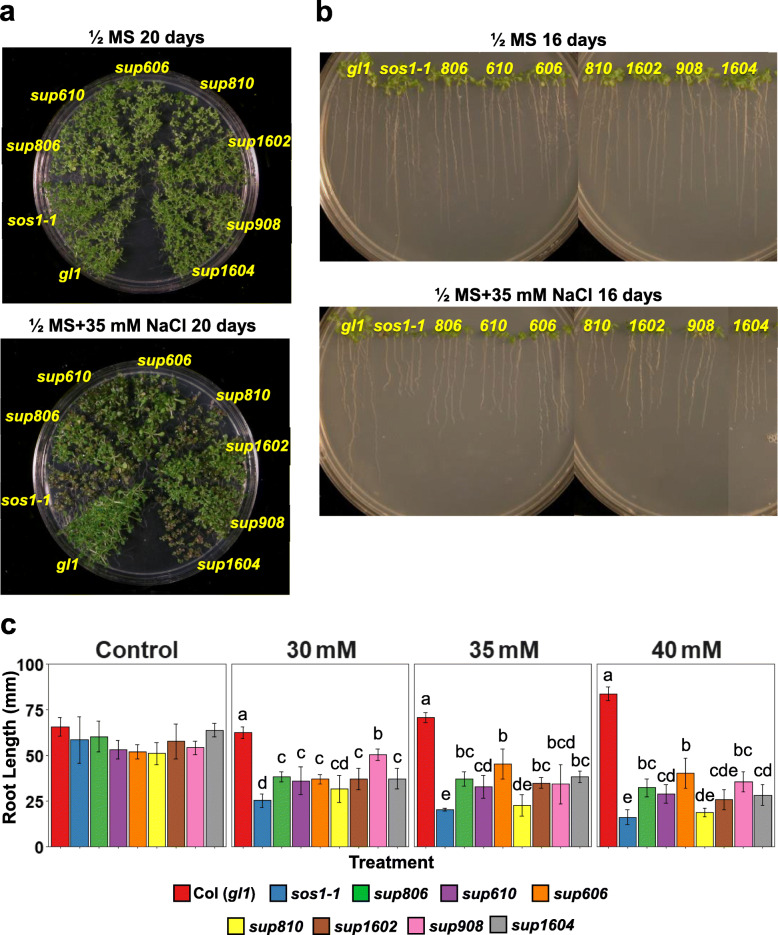
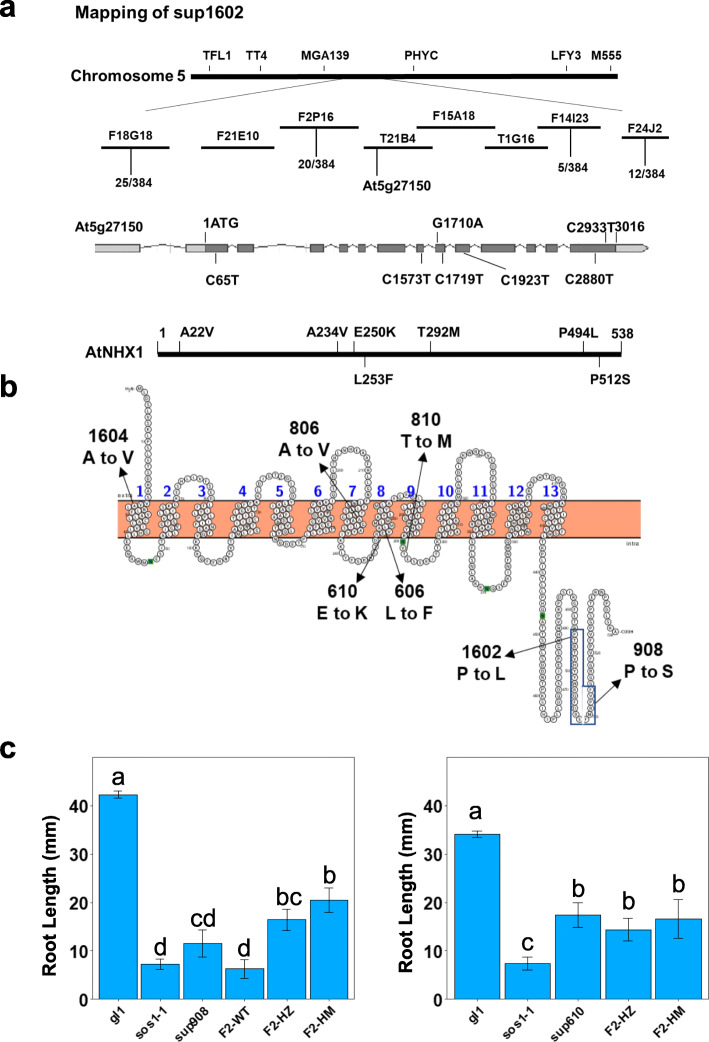
Gene mapping and sequencing revealed a group of sup mutants (sup606, 610, 806, 810, 908, 1602, and 1604) harboring mutations in the vacuolar Na+/H+ antiporter gene AtNHX1. These sup lines showed better growth in both shoots (Fig. 2a) and roots (Fig. 2b, c) compared to the sos1–1 mutant under salinity stress. The suppressor mutant sup1602 was initially chosen for positional cloning and the mutant gene was delimited to the region between the BAC clones F2P16 and F14I23 in chromosome 5 (Fig. 3a). After examining the genes within this region, the previously characterized salinity tolerance determinant gene AtNHX1 was found in this region and was an apparent candidate for the suppressor gene. Sequencing of the AtNHX1 gene in sup1602 mutant revealed a heterozygous C2880T nucleotide change in the coding region (Fig. 3a, Fig. S1), which suggested that the mutation could be a dominant, gain-of-function mutation. Subsequent sequencing of the AtNHX1 gene in all other suppressor lines identified the above-mentioned seven lines harboring mutations in AtNHX1 gene, among which four lines had homozygous mutations and three possess heterozygous mutations (Fig. 3a, b, Fig. S1). Crosses between the homozygous mutants sup908 and sup610 with wild-type Col (gl1) revealed that both heterozygotes and homozygotes of the suppressor lines displayed suppression of the sos1–1 salt sensitivity (Fig. 3c, Fig. S2), which further supported that the mutations in AtNHX1 gene are dominant mutations conferring salt tolerance. The suppressor mutations in the AtNHX1 gene resulted in amino acid substitutions in vital positions of the protein (Fig. 3b). The lines sup1604 and 806 possess an Ala to Val change in predicted transmembrane domains, while sup610 and sup606 have transmembrane residue changes of Glu to Lys and Leu to Phe, respectively. The sup810 mutant harbors an amino acid change of Tyr to Met in a loop, which alters a prospective N-glycosylation site (NVT to NVM). For sup1602 and 908, the mutations are situated in the C-terminal region of AtNHX1, which result in amino acid substitutions in the previously characterized calmodulin-interacting motif (Yamaguchi et al. 2005).
Fig. 2.
Phenotyping of the sup lines harboring mutations in AtNHX1. a The suppressor lines exhibit suppression of sos1 salt sensitivity in shoot growth under salinity stress (35 mM NaCl). b Root growth phenotype of the sup lines. c Quantification of the root length of WT, sos1–1 and the sup suppressor mutants. Bars represent means, and error bars represent standard errors (n = 5). Statistical significance among genotypes within treatments was computed with ANOVA and Tukey’s post-hoc HSD test (P < 0.05) and different statistical groups are represented by letters
Fig. 3.
Identification of the dominant mutations in AtNHX1 gene in the sup mutants. a Positional cloning of the sup1602 mutant gene. The sup mutant alleles were identified by sequencing the AtNHX1 gene in the sup mutants. Nucleotide changes and amino acid substitutions in the gene and protein of the sup mutants are shown. b Predicted secondary structure of AtNHX1 and the amino acid substitutions in the sup mutants. The structure was illustrated using Protter (Omasits et al. 2013). c The root lengths for the different genotypes indicating improved salinity tolerance in all genotypes carrying a sup AtNHX1 allele. HZ, heterozygote, HM, homozygote. Bars represent means, and error bars represent standard errors (n = 5). Statistical significance was computed with ANOVA and Tukey’s post-hoc HSD test (P < 0.05) and different statistical groups are represented by letters
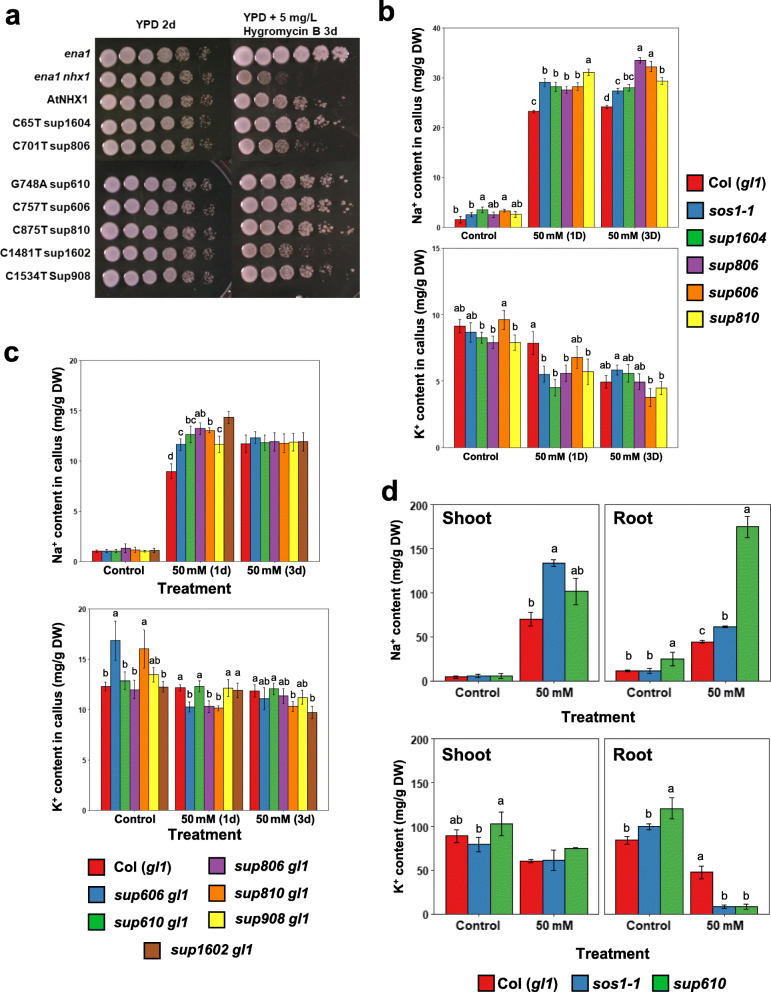
Gain-of-function mutations of AtNHX1 in the suppressor mutants were further verified by heterologous expression of the mutated AtNHX1 in yeast cells (Fig. 4a). As shown in previous studies (Gaxiola et al. 1999; Hernández et al. 2009), expression of the wild type AtNHX1 complemented and rescued the hygromycin sensitivity of the yeast mutant with a defective tonoplast Na+/H+ antiporter ScNHX1. Compared with the wild type AtNHX1, expression of the AtNHX1 suppressor variants sup1604, 610, 606, and 810 in yeast exhibited superior resistance to hygromycin (Fig. 4a), indicating increased activity of the variant transporters. Notably, sup806, 1602, and 908 did not show increased activity in the yeast cells, suggesting that the mechanisms of these alterations enhancing AtNHX1 function in plant may not exist in yeast. This is especially true for sup1602 and 908 which harbor mutations in the regulatory C-terminal region of the protein known to interact with and be inhibited by binding with AtCaM15 (Yamaguchi et al. 2005). This regulatory mechanism might be unique to plants or at least absent in yeast. Our findings also suggest that the suppressor mutations improve the efficacy of the AtNHX1 transporter through different mechanisms. In addition, genetic analysis using the T-DNA knockout mutant of AtNHX1 further supported the gain-of-function feature of the suppressor mutations. The double mutant sos1nhx1 displayed more salt sensitivity than the single mutant sos1 (Fig. S3), which is opposite to the enhanced salt tolerance of sos1 by the gain-of-function suppressor mutations (Fig. 2).
Fig. 4.
The suppressor mutations in AtNHX1 are gain-of-function mutations. a Yeast complementation assay showing enhanced activity of AtNHX1 carrying the sup mutations. AtNHX1 activity was assessed in yeast (Saccharomyces cerevisiae) by its capacity conferring tolerance to hygromycin B (5 mg L− 1). b, c Na+ and K+ contents in the callus cells of sup mutants in the sos1–1 background (b) and Col (gl1) background (c). Calli were sampled (n = 3) at one (1d) and 3 days (3d) after transferring to the agar medium containing 50 mM NaCl. Confidence intervals (CI) were used to determine statistically significant differences within treatments and letters denote different groups (P < 0.05). Bars represent means, and error bars represent the CI (n = 3). d Na+ and K+ contents in the shoots and roots of Col (gl1), sos1–1, and a representative sup mutant line, sup610. Bars represent means, and error bars represent standard errors (n ≥ 3). Statistical differences among genotypes within a treatment group was computed with ANOVA and Tukey’s post-hoc HSD test (P < 0.05, n ≥ 3) and different statistical groups are represented by letters
In accordance with the gain-of-function mutations in AtNHX1 in the sup mutants, the callus cells of the sup lines accumulated more Na+ than the sos1–1 mutant after 50 mM NaCl treatment for 3 days (Fig. 4b), which is likely due to enhanced AtNHX1 activity and thus increased Na+ sequestration into the vacuoles of the sup lines. The K+ contents in sos1–1 and sup lines were lower than the wild type after salt treatment for 1 day, perhaps mainly due to the effects of sos1 mutation on K+ uptake. Enhanced Na+ accumulation by the gain-of-function mutations in AtNHX1 was further confirmed by the analysis of Na+ and K+ in the callus cells of the AtNHX1 gain-of-function mutants in the wild type (Col (gl1)) background (Fig. 4c). The results showed that all the gain-of-function mutants accumulated significantly higher Na+ in the callus cells than the wild type after salt treatment for 1 day. However, the Na+ contents in all lines are comparable after 3 days salt treatment, suggesting a limit in the capacity for vacuolar Na+ sequestration (Fig. 4c).
Measurements of ion contents at the whole plant level provide insight to the possible mechanism of how the AtNHX1 variants suppress sos1 salt sensitivity (Fig. 4d). Under salinity stress, the sos1–1 mutant shoot accumulated higher Na+ than wild type, while the suppressor mutant sup610 had reduced Na+ accumulation compared with sos1–1 mutant. In the roots, sup610 showed the highest Na+ content among the three lines, while sos1–1 had significantly higher Na+ than the wild type under salt stress condition. These results suggest that the enhanced AtNHX1 activity in sup610 helps prevent the translocation of Na+ into the photosynthetic shoot tissues by accumulation of Na+ in the roots, likely through the sequestration of Na+ into the vacuoles of the root cells, thus reducing the rate of Na+ movement from root to shoot. The K+ content in shoot is comparable among the three lines, while both sos1–1 and sup610 displayed drastically decreased K+ accumulation in roots when compared with the wild type.
Mutations in the AtNHX1 C-terminal region disrupts the inhibitory binding site for AtCaM15
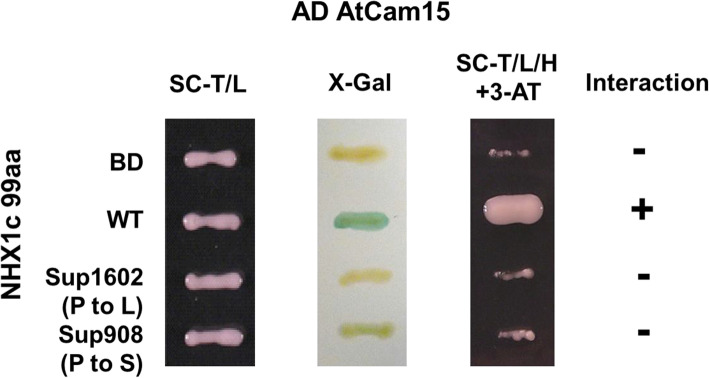
AtCaM15, a calmodulin-like protein, has been shown to inhibit the activity of AtNHX1 by binding to its C-terminal region in a Ca2+- and pH-dependent manner (Yamaguchi et al. 2005). Among the seven identified suppressor lines, two sup mutants, sup1602 and sup908, harbor amino acid substitutions in the regulatory C-terminal domain (Fig. 3b). A yeast two-hybrid interaction assay was conducted to determine the binding activity of AtCaM15 to these AtNHX1 variants (Fig. 5). As expected, the wild type AtNHX1 C-terminal region (99 amino acids) interacted with AtCaM15 as indicated in the X-Gal transactivation test and synthetic deficient media growth test. In contrast, the gain-of-function AtNHX1 variants sup1602 and 908 could not interact with AtCaM15. This result clearly indicates the disruption of the CaM-interacting domain in these two AtNHX1 variants. Since binding of AtCaM15 suppresses the transport activity of AtNHX1, disruption of the interaction in these two sup lines releases the inhibitory activity of AtCaM1 and enhances Na+ transport activity across the vacuolar membrane. We further tested whether a loss-of-function mutation in AtCaM15 could also suppress sos1 salt sensitivity since the gene knockout of AtCaM15 is supposed to eliminate its binding to and inhibition of AtNHX1. However, the sos1cam15 double mutant did not show enhanced salt tolerance but rather exhibited even slightly increased salt sensitivity (at 40 mM NaCl) compared to the sos1 single mutant (Fig. S4), These results suggest that, while AtCaM15 is important in the regulation of AtNHX1 activity, there may be other calmodulins or calmodulin-like proteins that have overlapping function with AtCaM15 in regulating AtNHX1. Additionally, it also implies that AtCaM15 plays a role in salt tolerance independent of AtNHX1.
Fig. 5.
Protein interaction assay between AtCaM15 and the C-terminal region of the wild type and sup alleles of AtNHX1. Yeast two-hybrid assay was conducted to determine the interactions. The C-terminal (99 aa) region of AtNHX1 was used as the bait fused with the Gal4 binding domain, while AtCaM15 was fused with the Gal4 activation domain. Positive interaction (+) was indicated by the blue color resulting from the metabolism of X-gal and vigorous growth under reductive synthetic media (SC-T/L/H + 3-AT)
Mutations in AtHKT1 led to suppression of sos1 salt sensitivity
Previous studies have shown that mutations in the AtHKT1 gene suppress the salt sensitivity of sos mutants (Rus et al. 2004). We therefore sequenced the AtHKT1 gene, including the promoter region, in all other sup mutants and identified six lines (sup109, 112, 906, 1123, 1601, and 1609) harboring mutations in AtHKT1 gene (Fig. 6a; sup1609 not shown). Na+ and K+ content in callus cells showed that, after one-day salt treatment, the sos1–1 mutant accumulated more Na+ than the wild type and athkt1 T-DNA mutant clearly suppressed Na+ accumulation in the sos1–1 mutant. While the hkt1 single mutant did not show effects on Na+ accumulation, mutation in hkt1 alone clearly affected K+ uptake under both control and salinity stress conditions (Fig. 6b). These results indicate that AtHKT1 controls Na+ and perhaps also K+ uptake at the cellular level. The sos1 suppressor sup1123 identified in this study behaved similarly with the previously characterized sos1–1hkt1 mutant harboring a T-DNA insertion in the AtHKT1 gene (Rus et al. 2004; Baek et al. 2010; Fig. 6b), further supporting that these sup lines harbor knockout mutations in the AtHKT1 gene. The six lines represented four unique mutations in AtHKT1 (Fig. 6c). The sup109 and 112 mutants had a substitution of Gly to Ala at the 36th amino acid residue, sup1601 and 1609 had a substitution of Ser to Phe at the 278th residue, sup1123 had a premature stop codon at the 309th codon, and sup906 had a single nucleotide deletion causing frame shaft after the 403rd codon. The single amino acid substitutions (sup109,112, 1601, and 1609) resulting in the loss of function could provide valuable information for understanding of the structure-function relations in AtHKT1 transporter.
Fig. 6.
Identification of sup lines with loss-of-function mutations in AtHKT1. a sos1 suppression phenotype of the sup mutants harboring mutations in AtHKT1 gene. b Na+ and K+ contents in the callus cells of Col-gl1, hkt1, sos1–1, sos1hkt1 and sup1123. Calli were sampled (n = 3) at one (1d) and 3 days (3d) after transferring to the agar medium containing 50 mM NaCl. Confidence intervals were computed and used to determine different statistical groups within treatments, which are denoted by the letters above each bar representing the means (P < 0.05, n = 3). c Secondary structure of AtHKT1 and the amino acid substitutions in the sup mutants. The secondary structure was illustrated using Protter (Omasits et al. 2013)
AtHKT1 loss-of-function and AtNHX1 gain-of-function additively suppress sos1 salt sensitivity
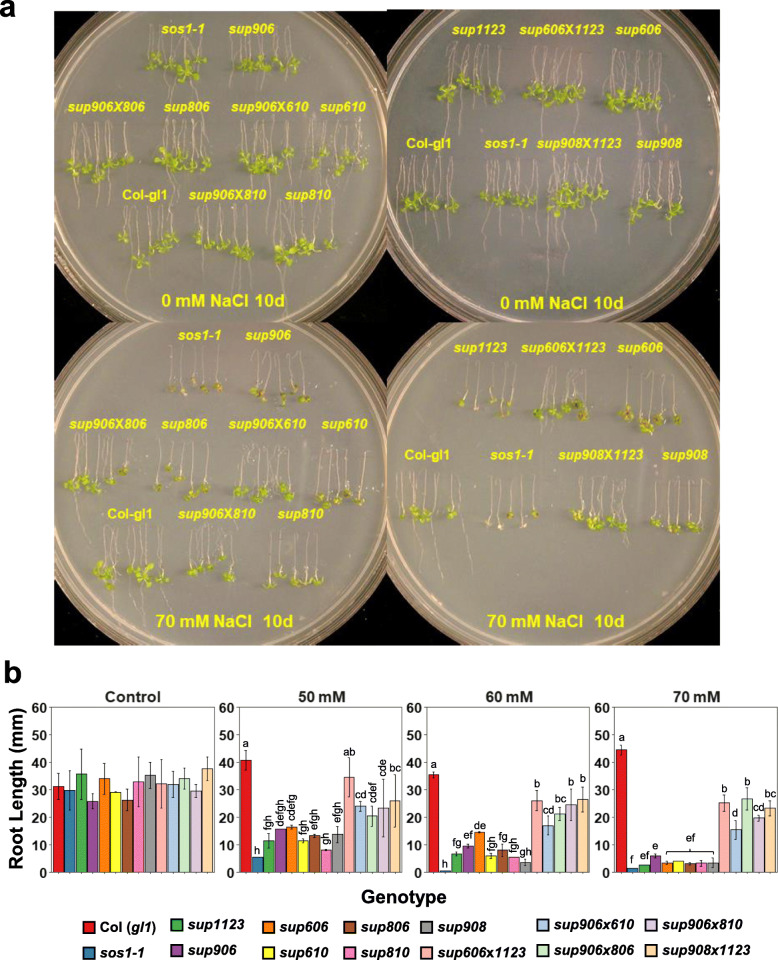
Since AtHKT1 controls the entry of Na+ into the cell and AtNHX1 controls the vacuolar influx of Na+, combining the loss-of-function mutation in AtHKT1 and gain-of-function mutation in AtNHX1 is expected to be additive in suppression of sos1 salt sensitivity. To test this hypothesis, combinations of different sup lines for AtHKT1 and AtNHX1 were genetically crossed, and double suppressor mutants were generated. Salt sensitivity assay showed that, at 70 mM NaCl, single sup mutants phenotypically resembled the sos1–1 mutant. In contrast, the double sup lines clearly showed better salt tolerance than the single sup lines in both root and shoot growth (Fig. 7a). Root length measurements showed a steady decrease in single sup lines along with increasing salt concentrations (Fig. 7b). At 70 mM NaCl, single sup lines did not show sos1 suppression phenotype in root growth. In contrast, double sup lines displayed stronger root growth compared to the tested single sup lines from 50 mM NaCl to 70 mM NaCl in addition to healthier shoots. These findings suggest a possibility of further improving plant salt tolerance by combining different salt tolerance mechanisms such as reducing cellular Na+ influx and increasing vacuolar Na+ sequestration to mitigate cytoplasmic Na+ toxicity.
Fig. 7.
Additive suppression effects of sup athkt1 and sup AtNHX1 mutants. a Root bending assay of wild type, sos1–1, sup athkt1, sup AtNHX1, and double suppressor mutants. The double suppressor mutants were generated by genetic crosses between the athkt1 and AtNHX1 sup lines. Five-day-old seedlings grown vertically in ½ MS agar (1.2%) medium were transferred to the same medium supplemented with different concentrations of NaCl, and the plates were put upside down for root bending assay. b Quantitative measurement of the root lengths of different genotypes in response to different concentrations of NaCl. Bars represent means, and error bars represent standard errors (n ≥ 3). Statistical significance among genotypes within a treatment group was computed with ANOVA and Tukey’s post-hoc HSD test (P < 0.05) and different statistical groups are represented by letters
Gain-of-function mutations in AtNHX1 improve salt tolerance in the wild type background
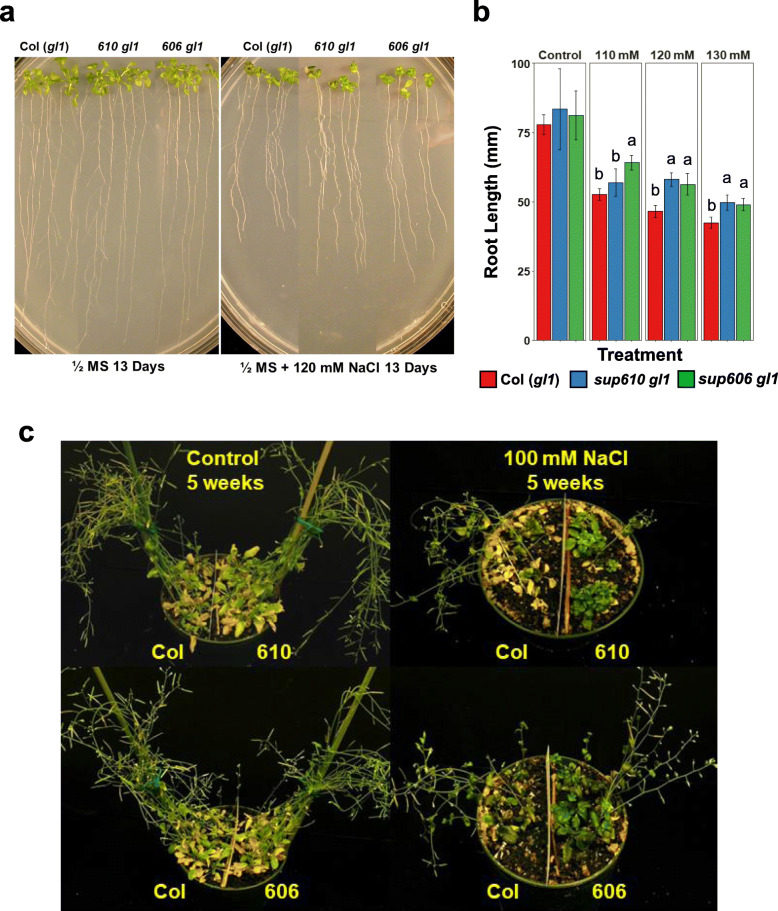
To determine the feasibility of using the identified AtNHX1 gain-of-function variants for improvement of salt tolerance in normal plant genotypes, AtNHX1 sup mutants were crossed with Col (gl1) and single AtNHX1 gain-of-function mutants without the sos1–1 mutation were identified from the resulting F2 populations. Under high salinity treatment (110 mM to 130 mM NaCl), plants harboring the sup610 or 606 AtNHX1 allele displayed slightly better root growth (Fig. 8a, b). These plants consistently showed significantly higher mean root lengths than Col (gl1) plants especially at 120 mM and 130 mM NaCl treatments. When plants were grown in soil to maturity under salt treatment (1/8 MS + 100 mM NaCl), the increased salt tolerance conferred by the AtNHX1 variant alleles was evident (Fig. 8c). Col (gl1) plants displayed more shoot wilting and bleached leaves, while lines with the sup606 and sup610 alleles exhibited far greener leaves under this treatment. However, the plants with the sup610 allele displayed delayed bolting compared to those with the sup606 allele under salt stress, which shows normal bolting and floral development like the wild type.
Fig. 8.
Gain-of-function AtNHX1 variants conferring increased salt tolerance in the wild-type background. a Root growth assay of the wild type Col (gl1) and the single AtNHX1 gain-of-function alleles 606 and 610 in Col (gl1) background. The single gain-of-function AtNHX1 mutant alleles were obtained by crossing the sup mutants with Col (gl1) background and determined by genotyping from the F2 population. b Quantitative measurement of the root lengths of Col (gl1) and the single gain-of-function mutants. Bars represent means, and error bars represent standard errors (n ≥ 3). Statistical significance among genotypes within a treatment group was computed with ANOVA and Tukey’s post-hoc HSD test (P < 0.05) and different statistical groups are represented by letters. c Soil-grown plants showing enhanced salt tolerance of the AtNHX1 gain-of-function mutant lines compared to the wild type
Discussion
Salinity stress is a widespread problem hampering crop productivity. Readjustment of cellular ion homeostasis is one of the crucial processes for plants to cope with salt stress (Zhu 2003; Basu et al. 2021). Salt tolerance is partly based on the capacity of the plant to compartmentalize toxic Na+ ions and thus reduce Na+ accumulation in the cytoplasm, where most of the cell’s metabolic processes occur (Munns and Tester 2008; Munns and Gilliham 2015). Therefore, enhancing the mechanisms that increase the capacity of plant cells to reduce cytoplasmic Na+ concentrations improves plant salinity tolerance. At a whole plant level, tissue-specific accumulation of Na+ is also an important mechanism to mitigate salt stress. For example, allelic variants of OsHKT1;1 in rice offer different capacities in excluding Na+ in roots (Campbell et al. 2017). In this study, we identified the genetic and physiological interactions between three membrane transporters, SOS1, HKT1, and NHX1 that are crucial for salt stress response and tolerance in Arabidopsis. We found that both loss-of-function of AtHKT1 and gain-of-function of AtNHX1 additively suppress sos1 salt sensitivity. Our findings indicate that these three transport systems act independently but coordinately to modulate cellular Na+ compartmentalization and plant salt stress response.
SOS1 is a vital plasma membrane Na+ transporter for salt exclusion by transporting Na+ out of the cell and thus preventing excess cytoplasmic Na+ accumulation in Arabidopsis (Shi et al. 2000; Shi et al. 2002). Its importance in maintaining ion homeostasis has been reported in other plant species (Olías et al. 2009; Razzaque et al. 2013; Feki et al. 2014; El Mahi et al. 2019; Wang et al. 2021). In this study, we found that two routes of lowering cellular Na+ accumulation counteract the loss of SOS1 function, i.e., prevention of cellular Na+ influx through AtHKT1 and enhancement of vacuolar Na+ compartmentalization by AtNHX1.
HKT1 was initially discovered to be a K+ influx transporter whose K+ transport activity was blocked and replaced with Na+ transport at high salinity conditions (Rubio et al. 1995). It has been hypothesized that at the whole plant level, HKT1 is important for lowering Na+ concentration in the xylem sap to prevent Na+ accumulation in photosynthetic tissues (Sunarpi et al. 2005). Thus, loss of this control for shoot Na+ accumulation leads to salt hypersensitivity of the hkt1 mutant. However, it has been shown that disruptive mutations of hkt1 can suppress sos1, sos2, and sos3 salt sensitivity (Rus et al. 2004). This implies that hkt1 is independent of the SOS pathway. This notion is also supported in our study, wherein the sup mutants carrying loss-of-function alleles of AtHKT1 improve the tolerance of sos1 mutant (Fig. 6). This may be simply due to the reduction of Na+ influx, leading to better survival under mild salt stress. Indeed, at higher Na+ concentrations and prolonged exposure to salt stress, these sup lines do not confer any advantage of salt tolerance. In addition, hkt1 mutations only confer salt tolerance in the sos1 background but not wild type background, suggesting a complex interaction between these two genes. Nonetheless, Baxter et al. (2010) showed that a weak allele of HKT1 in accessions growing in coastal regions confers increased salt tolerance. This further indicates that HKT1 is an evolutionarily important Na+ transporter in plant adaptation to saline environment and could be used as a marker for salt tolerance breeding in crops.
A previous study by Hernández et al. (2009) identified NHX1 mutants improving tolerance to ion toxicity in yeast by increasing its selectivity to K+ over Na+ and Li+. This suggests that gain-of-function mutations in NHX1 could improve salt tolerance. Through sos1 suppressor screening, we also identified a group of dominant and gain-of-function mutations in AtNHX1 conferring increased salt tolerance in both sos1 and wild type background (Figs. 2, 8, Fig. S3). These gain-of-function mutations could be classified into two types: 1) transmembrane domain mutants, and 2) regulatory domain mutants. The mutations in the transmembrane domains (TMs) of the three mutants (i.e., sup806, 610, 606) are concentrated around the predicted TM-7 and TM-8 regions and may directly affect the binding and transport of Na+ and H+. The sup1604 mutation is located in the predicted TM-1, which is further from TM-7 and TM-8 in the secondary structure. However, TM-1 might be a part of the catalytic region in the 3D structural context. Interestingly, both sup1604 and sup806 have an Ala to Val substitution, which is a subtle change in terms of amino acid properties but nonetheless increases its transport activity. This indicates that the length of the side chain of these two amino acids may play a role in Na+ and H+ transport by AtNHX1. Regardless, the enhanced activity of these variants was evidenced in the transformed yeast and in callus cells, especially for sup610 and 606 (Fig. 4a, c). The enhanced transport activity improves vacuolar Na+ sequestration, leading to better plant salt tolerance.
The mutation in sup810 is on a possible N-glycosylation site in the loop between TM-9 and TM-10 and thus may alter a regulatory mechanism. Protein glycosylation has been shown to be important for protein folding (Nagashima et al. 2018) and implicated in plant development and stress response (Trinidad et al. 2020). While the glycosylation of AtNHX1 at this site is unknown, the sup810 mutation increases AtNHX1 activity as evidenced by the yeast complementation assay (Fig. 4a). This suggests that glycosylation, if present at this site in AtNHX1, is a common inhibitory mechanism in plant and yeast. Glycosylation might block the ion binding and transport site of AtNHX1 since the NVT site is at the TMs possibly forming its ion transport channel (Fig. 3b). On the other hand, the mutations in sup908 and 1602 are on the C-terminal tail that has been shown to bind with and inhibited by AtCaM15 in a pH-dependent manner (Yamaguchi et al. 2005). Binding of AtCaM15 leads to a decrease in the Na+ transporter activity of AtNHX1, while maintaining its K+ transporter activity. It was hypothesized that this interaction represses AtNHX1 activity under normal growth conditions, and dissociation activates AtNHX1 under salt stress. Although this inhibitory mechanism may have some importance under ideal growing conditions, such an inhibition is clearly inadequate when vacuolar Na+ sequestration is required to reduce cytoplasmic Na+ under salinity stress conditions. Thus, the mutant alleles of sup908 and 1602 which are unable to be inhibited by AtCaM15 have elevated activity to deal with salt stress. While the full extent of the trade-offs associated with this type of mutations is not yet known, the identified gain-of-function mutations may be more beneficial than overexpression in improving salt tolerance in plants because the AtNHX1 variants resemble the wild type AtNHX1 and do not carry the same metabolic burden as overexpression. This hypothesis is supported by our observation that the sup alleles driven by the constitutive overexpressing promoter (CaMV 35S), although providing enhanced salt tolerance in the sos1–1 mutant, did not suppress the salt sensitivity of sos1 to the same degree as the identified sup lines (Fig. S5). In a system such as a plant under salt stress, limited metabolic productivity is a major constraint for survival, especially from the processes that consume ATP (Wu et al. 2016; Pabuayon et al. 2021).
The results presented here indicate the independent action of AtSOS1, AtNHX1, and AtHKT1 in maintaining ion homeostasis under salinity stress. It has been proposed that these three transporters act in support of each other to minimize the accumulation of Na+ in photosynthetic or metabolically active tissues (Zhang and Shi 2013; Gong et al. 2020). Cellular Na+ exclusion provided by AtSOS1 may be the primary mechanism of maintaining Na+ homeostasis, which is supported by the extremely salt sensitive phenotype of the sos1 mutants (Shi et al. 2000). In addition, the activities of AtHKT1 and AtNHX1, which both transport Na+ to prevent cytosolic Na+ accumulation, play crucial roles in cellular Na+ homeostasis. At the whole plant level, the accumulation of Na+ in parenchyma cells via the activity of AtHKT1 lowers Na+ concentration in the root xylem sap, leading to reduced Na+ translocation to shoots. AtNHX1 further reduces salt stress damage by compartmentalizing Na+ to the vacuoles and reducing Na+ accumulation in shoots. Therefore, these three transporters independently while synergistically function in maintaining Na+ homeostasis at both cellular and whole plant levels. Similar strategies in maintaining ion homeostasis have been reported for other plant species, including halophytes (Horie et al. 2012; Zhang et al. 2017). This indicates the conservation of these mechanisms in salt stress response in different plant species, while variant transporters may partly account for the capability of salinity tolerance.
The identified gain-of-function mutations in AtNHX1 in this study are mostly conserved in other important crop species. The alignment of the NHX1 homologs in cotton (Gossypium hirsutum), rice (Oryza sativa), maize (Zea mays), and soybean (Glycine max) with AtNHX1 showed conservation in five of the seven identified amino acids (Fig. S6). For the mutation in sup806 (Ala to Val at the 227th residue), all the other species examined have a natural Val residue. The only mutated residue that had a strong difference in AtNHX1 compared to its homologs is the mutation in sup1604 (Ala to Val at the 22nd residue), as other species have a consensus Ser residue instead of an Ala residue in AtNHX1. This result implies the importance of the mutation sites in NHX1 function for Na+ transport. In addition, the conservation of these alleles points to the possible target sites of NHX1 transporter for gene editing in major crops. Recent advances in gene editing technologies, particularly CRISPR-Cas, have become increasing prevalent in creating superior crop genotypes (Arora and Narula 2017; Steinwand and Ronald 2020). The improvements may be geared towards abiotic stresses, such as drought, salt, or heat, or towards pathogen and disease resistance (Zaidi et al. 2018; Zafar et al. 2019). However, the approach requires a target gene and alleles from which the edits can be patterned to. The results from our study provide viable alleles of NHX1 that can be created by gene editing in major crops, as majority of the functional mutations identified are well-conserved in major crop species.
To conclude, we have identified loss-of-function alleles of AtHKT1 and gain-of-function alleles of AtNHX1 confer salt tolerance in sos1 mutant. Notably, the identified gain-of-function mutations in AtNHX1 are all single amino acid substitutions resulting in enhanced functionalities. These mutations provide direct target sites for gene editing of NHX1 transporters in crops for improved salt tolerance. Moreover, these mutations provide valuable information for better understanding of the structure-function relation of Na+/H+ antiporter and its activity regulation. Future studies could use protein structure modeling to uncover the mechanistical importance of the identified amino acids of AtNHX1 in Na+ and H+ transport, which will assist computational design of superior NHX1 transporter for genetic modifications and improvement of salt tolerance in crops.
Materials and methods
Plant materials, sos1 suppressor screening, and genetic mapping
The sos1–1 mutant seeds were treated with 0.3% ethyl methanesulfonate (EMS) for 15 h for mutagenesis and the M2 population of the mutagenized seeds were generated for sos1 suppressor (named as sup) screening (Kim et al. 2006). The M2 seedlings were grown in ½ Murashige and Skoog (MS) agar medium (Phytotechnology Laboratories, KS) containing 30 mM NaCl and the seedlings showing better growth than sos1–1 seedlings were selected as putative sos1 suppressor mutants. After rescreening, the suppressor mutants were further confirmed by sequence verification of the sos1–1 mutation in the suppressor mutants. For map-based cloning, the sos1 suppressor mutant sup1602 was crossed with the sos1 T-DNA knockout alleles (ET11830/GT7225) in the Landsberg ecotype background. The F1 plants were selfed and the sos1 suppressor mutants were selected from the segregating F2 population by the root-bending assay (Wu et al. 1996). The F2 recombinant lines were used to map the sos1 suppressor gene using Col/Landsberg erecta polymorphic markers. The sup1602 mutation was mapped between the markers CER451900 and CER 449133, and the AtNHX1 gene in this region was considered as the prime candidate gene. The AtNHX1 gene was sequenced in sup1602 and all other sup mutants to determine the suppressor mutations. The inheritance pattern of the suppressor phenotype was determined through the crossing of sup908 and 610 mutants with the sos1–1 genotype. F2 lines from the crosses were genotyped using cleaved amplified polymorphic sequence (CAPS) markers. The sup908 allele was identified through cleavage by Sau3AI, which does not cleave the wild type AtNHX1 allele. The sup610 allele was identified through non-cleavage by MnlI, which cleaves the wild type AtNHX1 allele.
Genetic interaction analysis of sos1, atnhx1, and atcam15 mutants
To determine the genetic interaction between the loss-of-function of AtNHX1 and SOS1, the nhx1 T-DNA line was crossed with the sos1–1 mutant and nhx1sos1–1 double mutant was identified by genotyping. The presence of the T-DNA insertion was determined by PCR using the primers 5′-gtttctcctaagtaccttgcttgg-3′ and 5′-caaaggtatgcctgtttcatcga-3′, together with the T-DNA left border primer (5′-cattttataataacgctgcggacatctac-3′) (Apse et al. 2003). The T-DNA line for AtCaM15 (SALK_143508C) was identified from the SALK sequence-indexed T-DNA insertion collections (Alonso et al. 2003). The flanking T-DNA insertion primers 5′-atgagctgcgacggaggca-3′ and 5′-tcaaccccaagcattatcaaacg-3′ were used together with the primer LBb1 (5′-gcgtggaccgcttgctgcaact-3′) for mutant identification and homozygosity analysis. The cam15 T-DNA mutant was crossed with sos1–1 mutant and cam15sos1–1 doble mutant was generated for genetic analysis.
Yeast complementation assay
The AtNHX1 open reading frame was amplified via PCR using the primers 5′-ccggaattcgcatgttggattctctagtgtcgaaactg-3′ and 5′-ccgctcgagtcaagccttactaagatcaggaggg-3′. The amplified fragment was inserted into EcoRI and XhoI sites of pENTR™1A (Invitrogen, Carlsbad, CA). Site-directed mutagenesis was conducted to generate the mutations in AtNHX1 corresponding to the identified mutations in the suppressor lines by using the primers shown in Table S1, following manufacturer’s instructions (Stratagene, La Jolla, CA). The AtNHX1 ORF was recombined into the yeast vector pAG426GPD-ccdB (Addgene) using the Gateway LR clonase II Enzyme Mix (Invitrogen, Carlsbad, CA). The constructs were transformed into the yeast strain ena1nhx1 via the lithium acetate/single-stranded carrier DNA/polyethylene glycol method (Gietz and Woods 2002). The yeast complementation assay was done as described by Yamaguchi et al. (2005).
Salt stress tolerance assay
Seeds of the mutants and wild-type Col (gl1) were sterilized for 15 min with 20% Clorox bleach plus 0.1% Triton X-100 solution and were washed at least five times with sterilized water. The seeds were stratified at 4 °C for 2 days and sown on ½ MS medium with 1.5% sucrose and 1.2% agar. The seeds were then germinated vertically at 23 °C under a long-day cycle (16 h daylight/8 h night). Five-day-old seedlings were transferred onto ½ MS medium containing the designated NaCl concentration, 1.5% sucrose and 1.2% agar. Root lengths were measured from at least three plants (n ≥ 3). For soil salinity stress testing, ten-day-old seedlings grown in ½ MS agar medium were transferred to 10 cm pots with soil. Seven days after transplanting, the plants were watered indirectly (from the bottom) with 1/8 MS medium containing 100 mM NaCl weekly.
Yeast two-hybrid interaction assay
Interaction assays were conducted as described in Yamaguchi et al. (2005) with modifications. Briefly, the DNA fragment corresponding to the 99-aa region at the C-terminal end of AtNHX1 (and sup1602/908 AtNHX1 alleles) was amplified and cloned to the bait vector pDEST-GBKT7. Meanwhile, wild type AtCaM15 cDNA was cloned to the prey vector pDEST-GADT7. The bait and prey constructs were transformed into Saccharomyces cerevisiae strain Y190 and grown in synthetic media without tryptophan and leucine (SC-T/L; control), and synthetic media without tryptophan, leucine, histidine and supplemented with 25 mM 3-AT (3-amino-1,2,4-triazole, Sigma; SC-T/L/H; selective). X-gal staining (LacZ reporter assay) was used to further confirm the positive interaction. Positive interaction was indicated by blue color in the X-gal staining assay and growth in the selective medium (Jiang et al. 2013).
Measurement of Na+ and K+ content
Callus formation was induced from the mutants and wild-type seeds following the method described in Wu et al. (1996). The calli were transferred to callus induction medium containing 50 mM NaCl for salt stress treatment. The calli were then collected and dried in an 80 °C oven for at least 2 days and weighed. For ion content measurement in plants, 7-day-old seedling grown in agar medium were transferred to a hydroponic culture system for salt treatment and collection of shoot and root samples (Baek et al. 2010). The shoot and root samples were dried at 80 °C for at least 2 days. The dried samples (n ≥ 3) were digested in 3 mL concentrated HNO3 overnight followed by digestion at 100 °C until the solution clarified. The sample solution was diluted by 2.5% nitric acid. Na+ and K+ contents were measured with an atomic absorption spectrometer (Model 2380; Perkin-Elmer, Norwalk, CT).
Statistical analysis, sequence alignments, and protein transmembrane structure visualization
Statistical analyses for the physiological parameters were conducted with R v4.0.4 using the ‘agricolae’ package (R Core Team 2020; de Mendiburu and de Mendiburu 2019). Tukey’s HSD post-hoc tests following analysis of variance (ANOVA) tests were performed to discern differences between different sample groups in each parameter tested. Confidence intervals were also computed in some datasets to provide evidence of statistical significance. Sequences for AtHKT1 and AtNHX1 homologs from different species were gathered from UniProt (The UniProt Consortium 2020). Protein transmembrane structure visualization was conducted through Protter (Omasits et al. 2013) and alignments were conducted using Clustal Omega (Sievers and Higgins 2014).
Supplementary Information
Additional file 1: Table S1. Primers used for site-directed mutagenesis of AtNHX1. Fig. S1. Sequencing chromatogram of the identified sup AtNHX1 mutants. The zygosity of the sup mutants were identified using their respective sequencing chromatograms. Arrows indicate the mutated nucleotides. Fig. S2. Genotype and phenotype segregation in the F2 of the genetic crosses between sos1–1 and the sup908 and 610 mutants, verifying the dominant inheritance pattern of the mutations. The inserts show the genotypes of the F2 seedlings identified with cleaved amplified polymorphic sequence (CAPS) markers. The sup908 allele is identified through its cleavage by Sau3AI, which is absent in the wild type, while the sup610 allele is identified through its non-cleavage by MnlI. Root lengths of individual seedlings are shown with color-coded bars according to their zygosity for their AtNHX1 allele. Fig. S3. Growth phenotype of the sos1nhx1 double mutant in response to salt stress. a Root growth assay of wild type Col (gl1), sos1–1 and sos1nhx1. The sos1nhx1 double mutant was generated by crossing sos1–1 with the T-DNA insertion atnhx1 mutant allele. b Quantitative measurement of root lengths of these three genotypes in response to different concentrations of NaCl. Bars represent means, and error bars represent standard errors (n ≥ 3). Statistical significance among genotypes within treatments was computed with ANOVA and Tukey’s post-hoc HSD test (P < 0.05) and different statistical groups are represented by letters. Fig. S4. Growth phenotype of sos1cam15, sup1602, and sup908 in response to salt stress. The sos1cam15 double mutant was obtained by crossing sos1–1 with the T-DNA insertion mutant cam15. The suppressor mutants sup1602 and 908 were used as controls showing suppression of sos1–1 salt sensitivity, while the cam15 mutation did not suppress sos1–1 salt sensitive phenotype. Fig. S5. Overexpression of the sup alleles showed less suppression of sos1–1 salt sensitivity than the native sup alleles. a Root growth assay of wild type, sos1–1, sup mutants and sup-OE seedlings. The sup-OE genotypes are the transgenic plants overexpressing the AtNHX1 alleles in the sos1–1 genetic background using the 35S promoter driven the ORF of the AtNHX1 alleles. AtNHX1 ORF cloning and site-directed mutagenesis to generate the sup AtNHX1 alleles were described in “Materials and Methods”. The ORFs of AtNHX1 alleles were recombined into the plant expression vector pEarleyGate 100 (Earley et al. 2006), and the constructs were transformed into sos1–1 mutant to generate sup-OE plants using the floral dip method (Clough and Bent 1998). Homozygous transgenic lines were selected for salt sensitivity test. b Quantitative measurement of the root lengths. Bars represent mean root lengths, and error bars represent standard errors (n = 5). Statistical significance among genotypes within a treatment group was computed with ANOVA and Tukey’s post-hoc HSD test (P < 0.05, n = 5) and different statistical groups are represented by letters. Fig. S6. Partial sequence alignment of NHX1 homologs in Arabidopsis and major crop species. NHX1 homologs in major crop species (maize, rice, soybean, and cotton) were aligned and compared to the Arabidopsis AtNHX1 protein. Residues wherein mutations were identified in the sup lines are highlighted. Residues in yellow are not conserved in the other crop species compared to Arabidopsis, while residues in green are conserved in all species shown.
Acknowledgements
We thank Ms. Qing Feng for the screening of sos1 suppressor mutants.
Authors’ contributions
H.S. conceived the project; I.C.P., J.J., H.Q., and J-S.C. performed the experiments; H.S., I.C.P. and J.J. analyzed the data; J.J., I.C.P. and H.S. wrote the manuscript. The author(s) read and approved the final manuscript.
Funding
This work was supported by the US Department of Agriculture [National Research Initiative project 2007–35100-18378 to H.S.].
Availability of data and materials
The materials used in this study will be available for research upon request.
Declarations
Competing interests
H.S. is a member of the editorial board but was not involved in the journal’s review or any decisions related to this submission.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Isaiah Catalino M. Pabuayon and Jiafu Jiang contributed equally to this work.
References
- Ali A, Maggio A, Bressan RA, Yun D-J. Role and functional differences of HKT1-type transporters in plants under salt stress. Int J Mol Sci. 2019;20(5):1059. doi: 10.3390/ijms20051059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 1999;285(5431):1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- Apse MP, Sottosanto JB, Blumwald E. Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J. 2003;36(2):229–239. doi: 10.1046/j.1365-313X.2003.01871.x. [DOI] [PubMed] [Google Scholar]
- Arora L, Narula A (2017) Gene editing and crop improvement using CRISPR-Cas9 system. Front Plant Sci 8(1932). 10.3389/fpls.2017.01932 [DOI] [PMC free article] [PubMed]
- Assaha DVM, Ueda A, Saneoka H, Al-Yahyai R, Yaish MW (2017) The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front Physiol 8(509). 10.3389/fphys.2017.00509 [DOI] [PMC free article] [PubMed]
- Baek D, Jiang J, Chung J-S, Wang B, Chen J, Xin Z, Shi H (2010) Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant and Cell Physiology 52 (1):149-161. 10.1093/pcp/pcq182 [DOI] [PubMed]
- Bassil E, Tajima H, Liang Y-C, M-a O, Ushijima K, Nakano R, Esumi T, Coku A, Belmonte M, Blumwald E. The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell. 2011;23(9):3482–3497. doi: 10.1105/tpc.111.089581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Kumar A, Benazir I, Kumar G. Reassessing the role of ion homeostasis for improving salinity tolerance in crop plants. Physiol Plant. 2021;171(4):502–519. doi: 10.1111/ppl.13112. [DOI] [PubMed] [Google Scholar]
- Baxter I, Brazelton JN, Yu D, Huang YS, Lahner B, Yakubova E, Li Y, Bergelson J, Borevitz JO, Nordborg M, Vitek O, Salt DE. A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet. 2010;6(11):e1001193. doi: 10.1371/journal.pgen.1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthomieu P, Conejero G, Nublat A, Brackenbury WJ, Lambert C, Savio C, Uozumi N, Oiki S, Yamada K, Cellier F, Gosti F, Simonneau T, Essah PA, Tester M, Very AA, Sentenac H, Casse F. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 2003;22(9):2004–2014. doi: 10.1093/emboj/cdg207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumwald E. Sodium transport and salt tolerance in plants. Curr Opin Cell Biol. 2000;12(4):431–434. doi: 10.1016/S0955-0674(00)00112-5. [DOI] [PubMed] [Google Scholar]
- Blumwald E, Aharon GS, Apse MP. Sodium transport in plant cells. Biochim Biophys Acta. 2000;1465(1–2):140–151. doi: 10.1016/S0005-2736(00)00135-8. [DOI] [PubMed] [Google Scholar]
- Brini F, Hanin M, Mezghani I, Berkowitz GA, Masmoudi K. Overexpression of wheat Na+/H+ antiporter TNHX1 and H+-pyrophosphatase TVP1 improve salt- and drought-stress tolerance in Arabidopsis thaliana plants. J Exp Bot. 2007;58(2):301–308. doi: 10.1093/jxb/erl251. [DOI] [PubMed] [Google Scholar]
- Busoms S, Paajanen P, Marburger S, Bray S, Huang X-Y, Poschenrieder C, Yant L, Salt DE. Fluctuating selection on migrant adaptive sodium transporter alleles in coastal Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2018;115(52):E12443–E12452. doi: 10.1073/pnas.1816964115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MT, Bandillo N, Al Shiblawi FRA, Sharma S, Liu K, Du Q, Schmitz AJ, Zhang C, Véry A-A, Lorenz AJ, Walia H (2017) Allelic variants of OsHKT1;1 underlie the divergence between indica and japonica subspecies of rice (Oryza sativa) for root sodium content. PLoS Genet 13(6):e1006823. 10.1371/journal.pgen.1006823 [DOI] [PMC free article] [PubMed]
- Chen H, An R, Tang J-H, Cui X-H, Hao F-S, Chen J, Wang X-C. Over-expression of a vacuolar Na+/H+ antiporter gene improves salt tolerance in an upland rice. Mol Breed. 2007;19(3):215–225. doi: 10.1007/s11032-006-9048-8. [DOI] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Counillon L, Pouyssegur J. The expanding family of eucaryotic Na+/H+ exchangers. J Biol Chem. 2000;275(1):1–4. doi: 10.1074/jbc.275.1.1. [DOI] [PubMed] [Google Scholar]
- Davenport RJ, Munoz-Mayor A, Jha D, Essah PA, Rus A, Tester M (2007) The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ 30(4):497–507. 10.1111/j.1365-3040.2007.01637.x [DOI] [PubMed]
- de Mendiburu F, de Mendiburu MF. Package ‘agricolae’. R package version:1.2–8. 2019. [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45(4):616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- El Mahi H, Pérez-Hormaeche J, De Luca A, Villalta I, Espartero J, Gámez-Arjona F, Fernández JL, Bundó M, Mendoza I, Mieulet D, Lalanne E, Lee S-Y, Yun D-J, Guiderdoni E, Aguilar M, Leidi EO, Pardo JM, Quintero FJ. A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice. Plant Physiol. 2019;180(2):1046–1065. doi: 10.1104/pp.19.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feki K, Quintero FJ, Khoudi H, Leidi EO, Masmoudi K, Pardo JM, Brini F. A constitutively active form of a durum wheat Na+/H+ antiporter SOS1 confers high salt tolerance to transgenic Arabidopsis. Plant Cell Rep. 2014;33(2):277–288. doi: 10.1007/s00299-013-1528-9. [DOI] [PubMed] [Google Scholar]
- Fukada-Tanaka S, Inagaki Y, Yamaguchi T, Saito N, Iida S. Colour-enhancing protein in blue petals. Nature. 2000;407(6804):581. doi: 10.1038/35036683. [DOI] [PubMed] [Google Scholar]
- Gaxiola RA, Li J, Undurraga S, Dang LM, Allen GJ, Alper SL, Fink GR. Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc Natl Acad Sci U S A. 2001;98(20):11444–11449. doi: 10.1073/pnas.191389398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, Fink GR. The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc Natl Acad Sci U S A. 1999;96(4):1480–1485. doi: 10.1073/pnas.96.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/S0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Gong Z, Xiong L, Shi H, Yang S, Herrera-Estrella LR, Xu G, Chao D-Y, Li J, Wang P-Y, Qin F. Plant abiotic stress response and nutrient use efficiency. Sci China Life Sci. 2020;63(5):635–674. doi: 10.1007/s11427-020-1683-x. [DOI] [PubMed] [Google Scholar]
- Hamamoto S, Horie T, Hauser F, Deinlein U, Schroeder JI, Uozumi N. HKT transporters mediate salt stress resistance in plants: from structure and function to the field. Curr Opin Biotechnol. 2015;32:113–120. doi: 10.1016/j.copbio.2014.11.025. [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol. 2000;51(1):463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- Hernández A, Jiang X, Cubero B, Nieto PM, Bressan RA, Hasegawa PM, Pardo JM. Mutants of the Arabidopsis thaliana cation/H+ antiporter AtNHX1 conferring increased salt tolerance in yeast: the endosome/prevacuolar compartment is a target for salt toxicity. J Biol Chem. 2009;284(21):14276–14285. doi: 10.1074/jbc.M806203200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Karahara I, Katsuhara M. Salinity tolerance mechanisms in glycophytes: an overview with the central focus on rice plants. Rice. 2012;5(1):11. doi: 10.1186/1939-8433-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia ZP, McCullough N, Martel R, Hemmingsen S, Young PG. Gene amplification at a locus encoding a putative Na+/H+ antiporter confers sodium and lithium tolerance in fission yeast. EMBO J. 1992;11(4):1631–1640. doi: 10.1002/j.1460-2075.1992.tb05209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Wang B, Shen Y, Wang H, Feng Q, Shi H. The Arabidopsis RNA binding protein with K homology motifs, SHINY1, interacts with the C-terminal domain phosphatase-like 1 (CPL1) to repress stress-inducible gene expression. PLoS Genet. 2013;9(7):e1003625. doi: 10.1371/journal.pgen.1003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Zhu J, Kim K, Agarwal M, Fu X, Huang A, Zhu J-K. The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc Natl Acad Sci U S A. 2006;103(49):18816–18821. doi: 10.1073/pnas.0604711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Schumaker KS, Zhu JK. EMS mutagenesis of Arabidopsis. Methods Mol Biol. 2006;323:101–103. doi: 10.1385/1-59745-003-0:101. [DOI] [PubMed] [Google Scholar]
- Li J, Yang H, Peer WA, Richter G, Blakeslee J, Bandyopadhyay A, Titapiwantakun B, Undurraga S, Khodakovskaya M, Richards EL, Krizek B, Murphy AS, Gilroy S, Gaxiola R. Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science. 2005;310(5745):121–125. doi: 10.1126/science.1115711. [DOI] [PubMed] [Google Scholar]
- Maser P, Eckelman B, Vaidyanathan R, Horie T, Fairbairn DJ, Kubo M, Yamagami M, Yamaguchi K, Nishimura M, Uozumi N, Robertson W, Sussman MR, Schroeder JI. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 2002;531(2):157–161. doi: 10.1016/S0014-5793(02)03488-9. [DOI] [PubMed] [Google Scholar]
- Møller IS, Gilliham M, Jha D, Mayo GM, Roy SJ, Coates JC, Haseloff J, Tester M. Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell. 2009;21(7):2163–2178. doi: 10.1105/tpc.108.064568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, Gilliham M. Salinity tolerance of crops – what is the cost? New Phytol. 2015;208(3):668–673. doi: 10.1111/nph.13519. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59(1):651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Nagashima Y, von Schaewen A, Koiwa H. Function of N-glycosylation in plants. Plant Sci. 2018;274:70–79. doi: 10.1016/j.plantsci.2018.05.007. [DOI] [PubMed] [Google Scholar]
- Nguyen NT, Vu HT, Nguyen TT, Nguyen L-AT, Nguyen M-CD, Hoang KL, Nguyen KT, Quach TN (2019) Co-expression of Arabidopsis AtAVP1 and AtNHX1 to improve salt tolerance in soybean. Crop Sci 59(3):1133–1143. 10.2135/cropsci2018.10.0640
- Olías R, Eljakaoui Z, Pardo JM, Belver A. The Na+/H+ exchanger SOS1 controls extrusion and distribution of Na+ in tomato plants under salinity conditions. Plant Signal Behav. 2009;4(10):973–976. doi: 10.4161/psb.4.10.9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasits U, Ahrens CH, Müller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2013;30(6):884–886. doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
- Pabuayon ICM, Kitazumi A, Cushman KR, Singh RK, Gregorio GB, Dhatt B, Zabet-Moghaddam M, Walia H, de los Reyes BG (2021) Novel and transgressive salinity tolerance in recombinant inbred lines of rice created by physiological coupling-uncoupling and network rewiring effects. Front Plant Sci 12 (267). doi:10.3389/fpls.2021.615277 [DOI] [PMC free article] [PubMed]
- Park S, Li J, Pittman JK, Berkowitz GA, Yang H, Undurraga S, Morris J, Hirschi KD, Gaxiola RA. Up-regulation of a H+-pyrophosphatase (H+-PPase) as a strategy to engineer drought-resistant crop plants. Proc Natl Acad Sci U S A. 2005;102(52):18830–18835. doi: 10.1073/pnas.0509512102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior C, Potier S, Souciet JL, Sychrova H. Characterization of the NHA1 gene encoding a Na+/H+-antiporter of the yeast Saccharomyces cerevisiae. FEBS Lett. 1996;387(1):89–93. doi: 10.1016/0014-5793(96)00470-X. [DOI] [PubMed] [Google Scholar]
- Qiu Q-S. Plant endosomal NHX antiporters: activity and function. Plant Signal Behav. 2016;11(5):e1147643. doi: 10.1080/15592324.2016.1147643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci U S A 99(12):8436–8441. 10.1073/pnas.122224699 [DOI] [PMC free article] [PubMed]
- Quintero FJ, Blatt MR, Pardo JM. Functional conservation between yeast and plant endosomal Na+/H+ antiporters. FEBS Lett. 2000;471(2-3):224–228. doi: 10.1016/S0014-5793(00)01412-5. [DOI] [PubMed] [Google Scholar]
- Quintero FJ, Martinez-Atienza J, Villalta I, Jiang X, Kim W-Y, Ali Z, Fujii H, Mendoza I, Yun D-J, Zhu J-K, Pardo JM. Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc Natl Acad Sci U S A. 2011;108(6):2611–2616. doi: 10.1073/pnas.1018921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2020) R: a language and environment for statistical computing, Vienna
- Razzaque S, Elias SM, Biswas S, Haque T, Seraj ZI. Cloning of the plasma membrane sodium/hydrogen antiporter SOS1 for its over expression in rice. Plant Tissue Cult Biotechnol. 2013;23(2):263–273. doi: 10.3329/ptcb.v23i2.17527. [DOI] [Google Scholar]
- Reguera M, Bassil E, Tajima H, Wimmer M, Chanoca A, Otegui MS, Paris N, Blumwald E. pH regulation by NHX-type antiporters is required for receptor-mediated protein trafficking to the vacuole in Arabidopsis. Plant Cell. 2015;27(4):1200–1217. doi: 10.1105/tpc.114.135699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rosales MP, Galvez FJ, Huertas R, Aranda MN, Baghour M, Cagnac O, Venema K. Plant NHX cation/proton antiporters. Plant Signal Behav. 2009;4(4):265–276. doi: 10.4161/psb.4.4.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio F, Gassmann W, Schroeder JI. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science. 1995;270(5242):1660–1663. doi: 10.1126/science.270.5242.1660. [DOI] [PubMed] [Google Scholar]
- Rus A, Lee BH, Munoz-Mayor A, Sharkhuu A, Miura K, Zhu JK, Bressan RA, Hasegawa PM. AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol. 2004;136(1):2500–2511. doi: 10.1104/pp.104.042234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A, Yokoi S, Sharkhuu A, Reddy M, Lee BH, Matsumoto TK, Koiwa H, Zhu JK, Bressan RA, Hasegawa PM. AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc Natl Acad Sci U S A. 2001;98(24):14150–14155. doi: 10.1073/pnas.241501798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo DP, Kumar S, Mishra S, Kobayashi Y, Panda SK, Sahoo L. Enhanced salinity tolerance in transgenic mungbean overexpressing Arabidopsis antiporter (NHX1) gene. Mol Breed. 2016;36(10):1–15. doi: 10.1007/s11032-016-0564-x. [DOI] [Google Scholar]
- Shen G, Wei J, Qiu X, Hu R, Kuppu S, Auld D, Blumwald E, Gaxiola R, Payton P, Zhang H. Co-overexpression of AVP1 and AtNHX1 in cotton further improves drought and salt tolerance in transgenic cotton plants. Plant Mol Biol Report. 2015;33(2):167–177. doi: 10.1007/s11105-014-0739-8. [DOI] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci U S A. 2000;97(12):6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Lee BH, Wu SJ, Zhu JK. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol. 2003;21(1):81–85. doi: 10.1038/nbt766. [DOI] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell. 2002;14(2):465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Zhu J-K. Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and abscisic acid. Plant Mol Biol. 2002;50(3):543–550. doi: 10.1023/A:1019859319617. [DOI] [PubMed] [Google Scholar]
- Sievers F, Higgins DG. Clustal omega, accurate alignment of very large numbers of sequences. Methods Mol Biol (Clifton, NJ) 2014;1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- Steinwand MA, Ronald PC. Crop biotechnology and the future of food. Nat Food. 2020;1(5):273–283. doi: 10.1038/s43016-020-0072-3. [DOI] [Google Scholar]
- Sunarpi HT, Motoda J, Kubo M, Yang H, Yoda K, Horie R, Chan WY, Leung HY, Hattori K, Konomi M, Osumi M, Yamagami M, Schroeder JI, Uozumi N. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J. 2005;44(6):928–938. doi: 10.1111/j.1365-313X.2005.02595.x. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2020;49(D1):D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinidad JL, Pabuayon ICM, Kohli A (2020) Harnessing protein posttranslational modifications for plant improvement. In: Tuteja N, Tuteja R, Passricha N, Saifi SK (eds) Advancement in crop improvement techniques. Woodhead Publishing, Cambridge. pp 385–401 10.1016/B978-0-12-818581-0.00023-1
- Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JI. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae 1. Plant Physiol. 2000;122(4):1249–1260. doi: 10.1104/pp.122.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hong Y, Li Y, Shi H, Yao J, Liu X, Wang F, Huang S, Zhu G, Zhu J-K. Natural variations in SlSOS1 contribute to the loss of salt tolerance during tomato domestication. Plant Biotechnol J. 2021;19(1):20–22. doi: 10.1111/pbi.13443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Yan Q, Jones JA, Tang YJ, Fong SS, Koffas MAG (2016) Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol 34(8):652–664 10.1016/j.tibtech.2016.02.010 [DOI] [PubMed]
- Wu SJ, Ding L, Zhu JK. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell. 1996;8(4):617–627. doi: 10.1105/tpc.8.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Aharon GS, Sottosanto JB, Blumwald E. Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2+- and pH-dependent manner. Proc Natl Acad Sci U S A. 2005;102(44):16107–16112. doi: 10.1073/pnas.0504437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Fukada-Tanaka S, Inagaki Y, Saito N, Yonekura-Sakakibara K, Tanaka Y, Kusumi T, Iida S. Genes encoding the vacuolar Na+/H+ exchanger and flower coloration. Plant Cell Physiol. 2001;42(5):451–461. doi: 10.1093/pcp/pce080. [DOI] [PubMed] [Google Scholar]
- Zafar SA, Zaidi SS-e-A, Gaba Y, Singla-Pareek SL, Dhankher OP, Li X, Mansoor S, Pareek A. Engineering abiotic stress tolerance via CRISPR/ Cas-mediated genome editing. J Exp Bot. 2019;71(2):470–479. doi: 10.1093/jxb/erz476. [DOI] [PubMed] [Google Scholar]
- Zaidi SS-e-A, Mukhtar MS, Mansoor S. Genome editing: targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 2018;36(9):898–906. doi: 10.1016/j.tibtech.2018.04.005. [DOI] [PubMed] [Google Scholar]
- Zhang HX, Blumwald E. Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol. 2001;19(8):765–768. doi: 10.1038/90824. [DOI] [PubMed] [Google Scholar]
- Zhang J-L, Shi H. Physiological and molecular mechanisms of plant salt tolerance. Photosynth Res. 2013;115(1):1–22. doi: 10.1007/s11120-013-9813-6. [DOI] [PubMed] [Google Scholar]
- Zhang W-D, Wang P, Bao Z, Ma Q, Duan L-J, Bao A-K, Zhang J-L, Wang S-M (2017) SOS1, HKT1;5, and NHX1 synergistically modulate Na+ homeostasis in the halophytic grass Puccinellia tenuiflora. Front Plant Sci 8(576). 10.3389/fpls.2017.00576 [DOI] [PMC free article] [PubMed]
- Zhu JK. Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003;6(5):441–445. doi: 10.1016/S1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Primers used for site-directed mutagenesis of AtNHX1. Fig. S1. Sequencing chromatogram of the identified sup AtNHX1 mutants. The zygosity of the sup mutants were identified using their respective sequencing chromatograms. Arrows indicate the mutated nucleotides. Fig. S2. Genotype and phenotype segregation in the F2 of the genetic crosses between sos1–1 and the sup908 and 610 mutants, verifying the dominant inheritance pattern of the mutations. The inserts show the genotypes of the F2 seedlings identified with cleaved amplified polymorphic sequence (CAPS) markers. The sup908 allele is identified through its cleavage by Sau3AI, which is absent in the wild type, while the sup610 allele is identified through its non-cleavage by MnlI. Root lengths of individual seedlings are shown with color-coded bars according to their zygosity for their AtNHX1 allele. Fig. S3. Growth phenotype of the sos1nhx1 double mutant in response to salt stress. a Root growth assay of wild type Col (gl1), sos1–1 and sos1nhx1. The sos1nhx1 double mutant was generated by crossing sos1–1 with the T-DNA insertion atnhx1 mutant allele. b Quantitative measurement of root lengths of these three genotypes in response to different concentrations of NaCl. Bars represent means, and error bars represent standard errors (n ≥ 3). Statistical significance among genotypes within treatments was computed with ANOVA and Tukey’s post-hoc HSD test (P < 0.05) and different statistical groups are represented by letters. Fig. S4. Growth phenotype of sos1cam15, sup1602, and sup908 in response to salt stress. The sos1cam15 double mutant was obtained by crossing sos1–1 with the T-DNA insertion mutant cam15. The suppressor mutants sup1602 and 908 were used as controls showing suppression of sos1–1 salt sensitivity, while the cam15 mutation did not suppress sos1–1 salt sensitive phenotype. Fig. S5. Overexpression of the sup alleles showed less suppression of sos1–1 salt sensitivity than the native sup alleles. a Root growth assay of wild type, sos1–1, sup mutants and sup-OE seedlings. The sup-OE genotypes are the transgenic plants overexpressing the AtNHX1 alleles in the sos1–1 genetic background using the 35S promoter driven the ORF of the AtNHX1 alleles. AtNHX1 ORF cloning and site-directed mutagenesis to generate the sup AtNHX1 alleles were described in “Materials and Methods”. The ORFs of AtNHX1 alleles were recombined into the plant expression vector pEarleyGate 100 (Earley et al. 2006), and the constructs were transformed into sos1–1 mutant to generate sup-OE plants using the floral dip method (Clough and Bent 1998). Homozygous transgenic lines were selected for salt sensitivity test. b Quantitative measurement of the root lengths. Bars represent mean root lengths, and error bars represent standard errors (n = 5). Statistical significance among genotypes within a treatment group was computed with ANOVA and Tukey’s post-hoc HSD test (P < 0.05, n = 5) and different statistical groups are represented by letters. Fig. S6. Partial sequence alignment of NHX1 homologs in Arabidopsis and major crop species. NHX1 homologs in major crop species (maize, rice, soybean, and cotton) were aligned and compared to the Arabidopsis AtNHX1 protein. Residues wherein mutations were identified in the sup lines are highlighted. Residues in yellow are not conserved in the other crop species compared to Arabidopsis, while residues in green are conserved in all species shown.
Data Availability Statement
The materials used in this study will be available for research upon request.