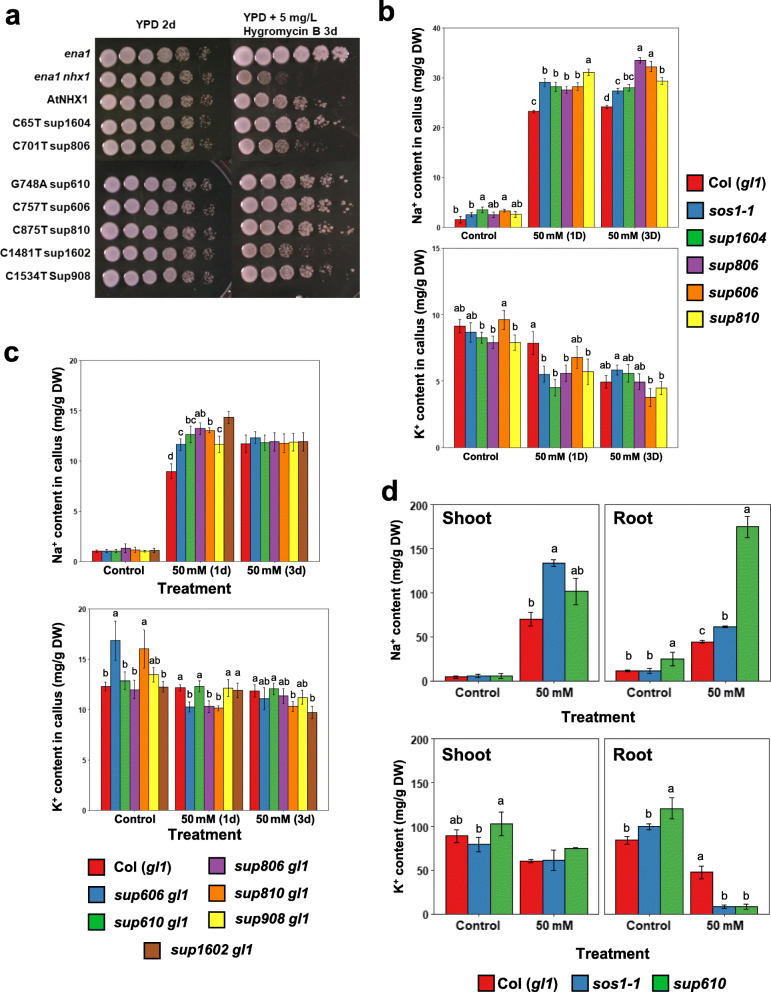
Fig. 4.
The suppressor mutations in AtNHX1 are gain-of-function mutations. a Yeast complementation assay showing enhanced activity of AtNHX1 carrying the sup mutations. AtNHX1 activity was assessed in yeast (Saccharomyces cerevisiae) by its capacity conferring tolerance to hygromycin B (5 mg L− 1). b, c Na+ and K+ contents in the callus cells of sup mutants in the sos1–1 background (b) and Col (gl1) background (c). Calli were sampled (n = 3) at one (1d) and 3 days (3d) after transferring to the agar medium containing 50 mM NaCl. Confidence intervals (CI) were used to determine statistically significant differences within treatments and letters denote different groups (P < 0.05). Bars represent means, and error bars represent the CI (n = 3). d Na+ and K+ contents in the shoots and roots of Col (gl1), sos1–1, and a representative sup mutant line, sup610. Bars represent means, and error bars represent standard errors (n ≥ 3). Statistical differences among genotypes within a treatment group was computed with ANOVA and Tukey’s post-hoc HSD test (P < 0.05, n ≥ 3) and different statistical groups are represented by letters