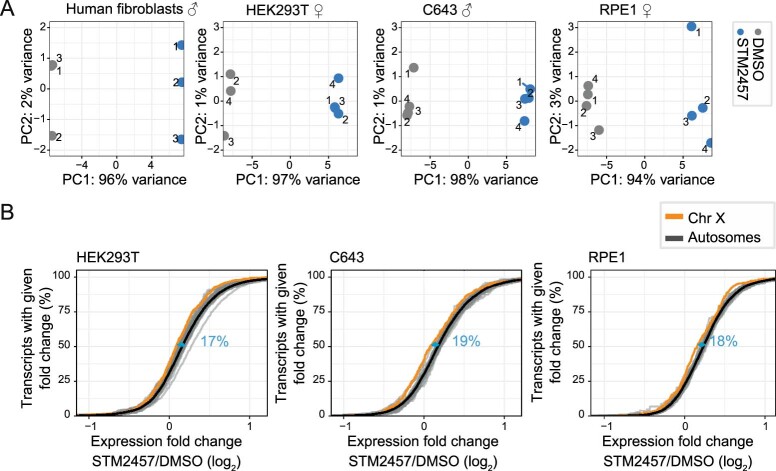
Extended Data Fig. 5. RNA-seq upon m6A depletion reveals upregulation of autosomal transcripts in human cell lines.
a. Principal component analyses for replicates of RNA-seq experiments under m6A-depleted and control conditions for human primary fibroblasts (STM2457, 9 h), HEK293T cells, C643 cells and RPE1 cells (STM2457, 24 h). Replicate number given next to each data point. b. X-chromosomal transcripts show significantly lower fold changes upon m6A depletion than autosomal transcripts (P value = 6.92e-06 [HEK293T, n = 12,856 of autosomal transcripts, n = 443 of X-chromosomal transcripts], P value = 4.53e-05 [C643, n = 11,109 of autosomal transcripts, n = 383 of X-chromosomal transcripts], P value = 0.0001901 [RPE1, n = 10,732 of autosomal transcripts, n = 347 of X-chromosomal transcripts], Wilcoxon rank-sum test). Cumulative fraction of transcripts on individual autosomes (grey) and the X chromosome (orange) that show a given fold change (log2) in m6A-depleted (STM2457, 24 h) over control conditions for HEK293T, C643, and RPE1 cells. Mean expression changes for all autosomes are shown as black line. Effect sizes (blue) shown the shift in medians, expressed as percent of the average IQR of autosomal and X-chromosomal genes (see Methods).