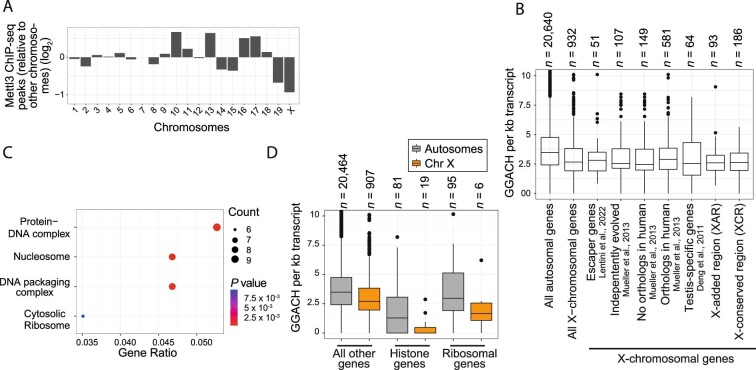
Extended Data Fig. 8. The number of GGACH motifs is reduced on transcripts encoding histones and ribosomal proteins.
a. The X chromosome harbours fewer Mettl3 ChIP-seq peaks. The number of published ChIP-seq peaks (normalised by chromosome length) per chromosome relative to peaks on all other chromosomes (log2). b. Different gene sets on the X-chromosome are similarly depleted in GGACH motifs. Quantification of GGACH motifs of all autosomal or X-chromosomal genes is compared to the following gene sets: escaper genes, independently evolved genes, genes with or without orthologs on the human X chromosome, testis-specific genes or genes residing in the X-added region (XAR) and X-conserved region (XCR). Numbers of genes are given in the figure (n). Boxes represent quartiles, centre lines denote medians, and whiskers extend to most extreme values within 1.5× interquartile range. c. X-chromosomal genes with low GGACH motif numbers are associated with DNA packaging or the cytosolic ribosome. Gene ontology (GO) enrichment analysis of the 200 genes with the lowest density of GGACH motifs on the X chromosome. P values were calculated by overrepresentation analysis (see Methods). d. Histone and ribosomal protein-encoding genes on the X chromosome are depleted in GGACH motifs. Quantification of GGACH motifs for histone-encoding and ribosomal protein-encoding genes on autosomes or on the X chromosome. Numbers of genes are given in the figure (n). Boxes as in b.