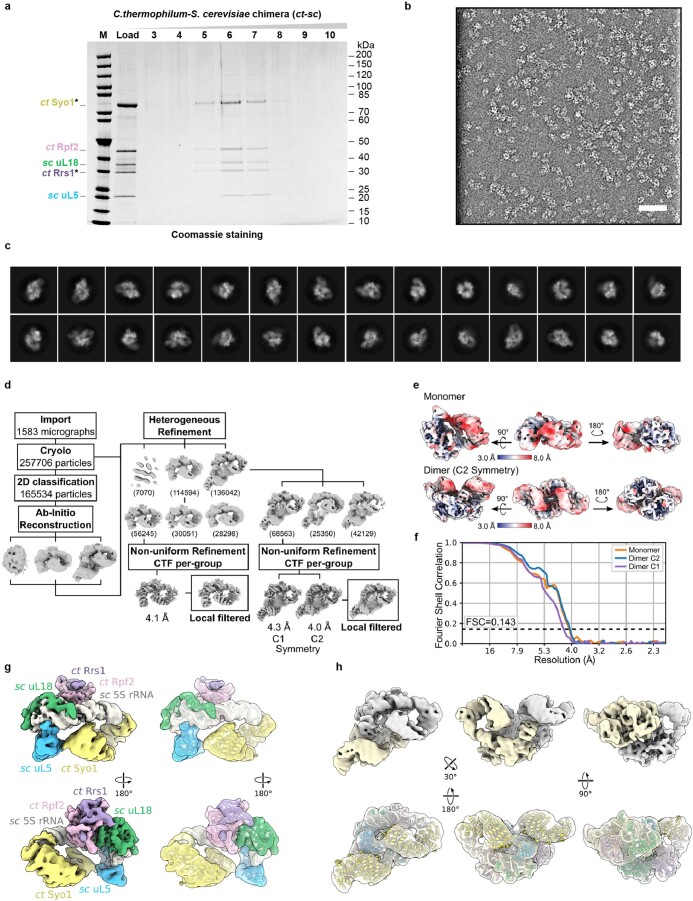
Extended Data Fig. 2. Purification, cryo-EM analysis and structure of the ct–sc 5S RNP chimera.
a, Split-tag affinity purification of the ct–sc 5S RNP chimera from yeast cells via ProtA-TEV-ctSyo1 and FLAG-ctRrs1 under conditions of ctRpf2 co-expression. The final FLAG eluate (Load) was separated by sucrose gradient centrifugation, and fractions were analyzed by SDS-PAGE. Bait proteins are labeled with asterisks. M: Molecular weight marker. Labeled bands were identified by MS. This purification has been done at least 5 times with similar results. b, c, Cryo-EM representative micrograph of the ct–sc 5S RNP chimera low-pass filtered to 20 Å, displayed with inverted contrast (out of 1583 micrographs used for processing) (b) and selected 2D classification averages (c). d, Cryo-EM processing scheme. The overall resolution of the final 3D reconstructions was 4.1 Å for the monomeric 5S RNP and 4.3 Å/4.0 Å for the dimeric 5S RNP without/with application of C2 symmetry. e, Final 3D reconstructions filtered and colored according to local resolutions. f, FSC curves of the final 3D reconstructions (masked). g, h, Cryo-EM densities and fitted models of the ct–sc 5S RNP assembled with C. thermophilum and S. cerevisiae factors for the monomeric (g) and dimeric (h) forms. 5S RNP factors are labeled in (g).