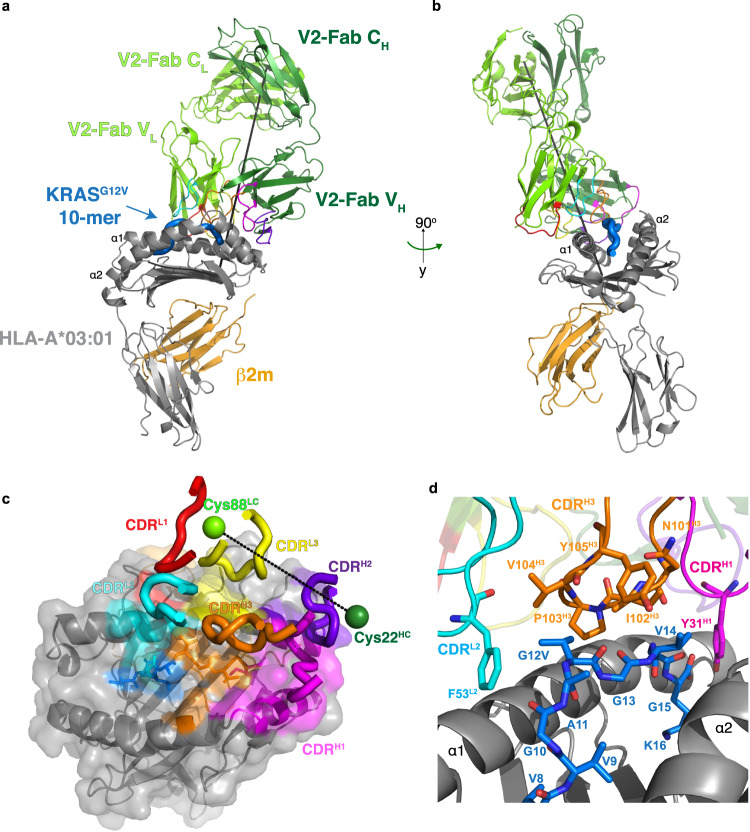
Fig. 3. Structural basis of recognition of V2-Fab.
a Overall structure of V2-Fab/KRASG12V-HLA-A*03:01 (PDB ID 7STF). The structure is colored according to Fig. 2. The CDRs are colored: L1 (red), L2 (cyan), L3 (yellow), H1 (magenta), H2 (purple), H3 (orange). The incident or ‘tilt’ angle is represented by a black line. b Structure of the V2-Fab/KRASG12V-HLA-A*03:01 complex shown in (a) at 90° rotation of that view. c Bird’s-eye view of the surface representation of the HLA-A*03:01 shown in gray, KRASG12V peptide shown in blue, and the contacting residues colored according to labeled CDRs of the V2-Fab, as in (a). The crossing angle, calculated from a vector between Cys22 from the V2 variable heavy chain and Cys88 from the V2 variable light chain, is shown as a dashed line. d Zoomed-in view of the KRASG12V peptide (aa Val8-Lys16) centered on the site of mutation, with interacting residues of CDRH3 (orange) and CDRL2 (cyan) shown as sticks.