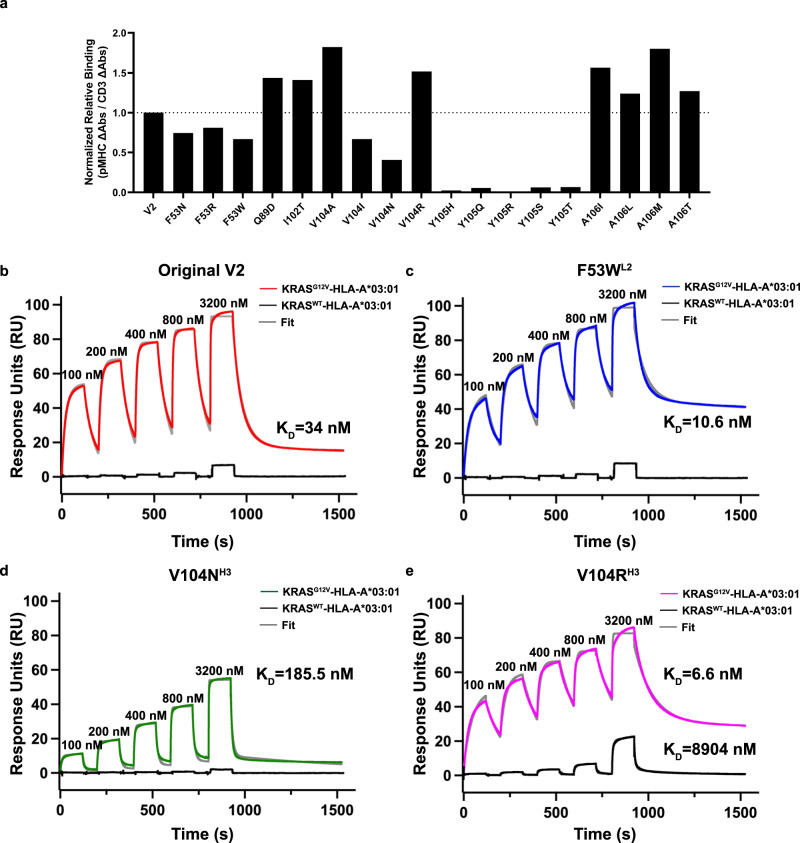
Fig. 6. Binding affinity and biophysical characterization of the V2 variant scDbs.
a scDbs were applied to ELISA plates coated with CD3ε/CD3δ heterodimer, KRASG12V-HLA-A*03:01, or the KRASG12WT-HLA-A*03:01 monomer. Relative binding was calculated as the ratio of KRASG12V–pHLA binding:CD3 binding for each scDb, and then reported as the percent of the original V2 scDb’s relative binding. n = 3 for each target. b scDb binding to KRASG12V-HLA-A*3:01 (red) and KRASWT-HLA-A*3:01 (black) was evaluated by single-cycle kinetics SPR. There was negligible binding to KRASWT–pHLA (black line). The data for KRASG12V-HLA-A*3:01 was fit with two state binding kinetics (gray line) for b original V2 scDb (KD = 34 nM), c F53WL2 scDb (KD = 10.6 nM), d V104NH3 scDb (KD = 185.5 nM) and e V104RH3 scDb (KD = 6.6 nM). V104RH3 binds to the KRASWT-pHLA with a KD > 3.2 µM. All sensorgrams are a representative experiment of n = 3 independent experiments.