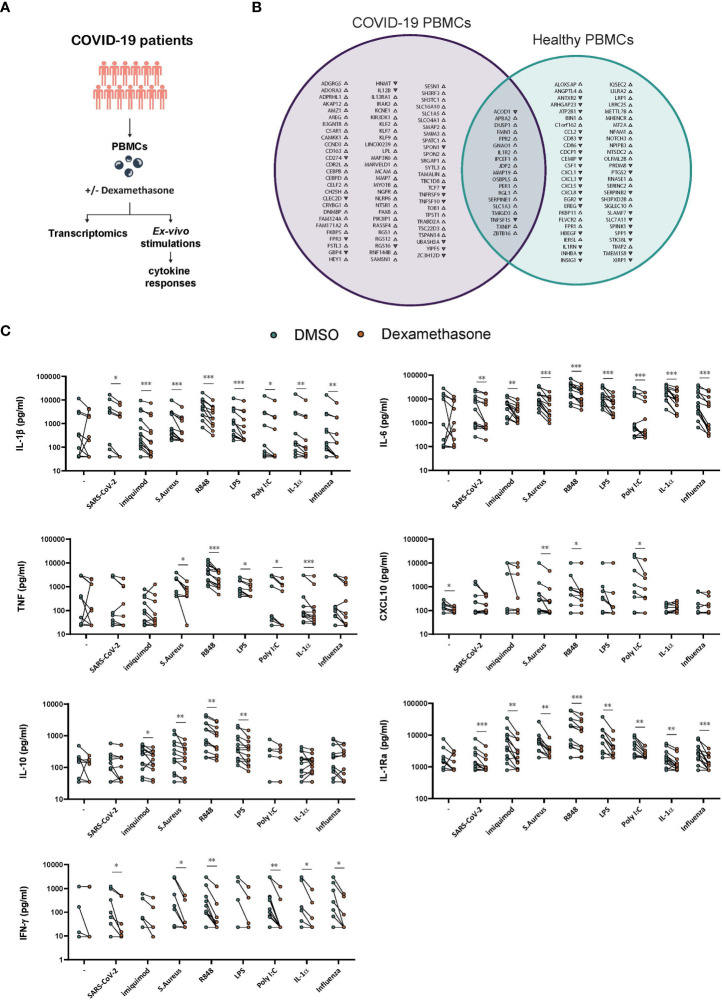
Figure 5.
Ex vivo effects of dexamethasone in stimulated PBMCs derived from COVID-19 patients. (A) Experimental setup for profiling the ex vivo transcriptome and cytokine responses of stimulated PBMCs derived from COVID-19 patients in the absence (DMSO) or presence of dexamethasone. (B) PBMCs derived from COVID-19 patients or healthy individuals were stimulated ex vivo with heat-inactivated SARS-CoV-2 for 4h in the absence (DMSO) or presence of dexamethasone and analyzed for their transcriptome profile. In the Venn diagram the genes significantly altered by dexamethasone only in PBMCs from COVID-19 patients are depicted with purple whereas the genes significantly altered by dexamethasone only in PBMCs from healthy individuals (as referred in Figure 1F ) are depicted with green. Gene upregulation or downregulation is indicated by the respective arrowheads. (n=5 COVID-19 patients, n=5 healthy individuals, FDR < 0.05). (C) PBMCs derived from COVID-19 patients were challenged ex vivo with heat-inactivated SARS-CoV-2, heat-inactivated influenza H1N1, heat-inactivated S. Aureus, the TLR ligands Imiquimod, R848, LPS or Poly I:C, the pro-inflammatory cytokine IL-1α, or left unstimulated for 24h in the absence (DMSO) or presence of dexamethasone (10nM), and the production of IL-1β, IL-6, TNF, CXCL10, IL-10 and IL-1Ra was assessed in supernatants after 24h (n=13 COVID-19 patients). The production of IFN-γ was assessed in the supernatants after 7d culture (n=12 COVID-19 patients). *: P value < 0.05, **: P value < 0.01, ***: P value < 0.001, Wilcoxon signed-rank test.