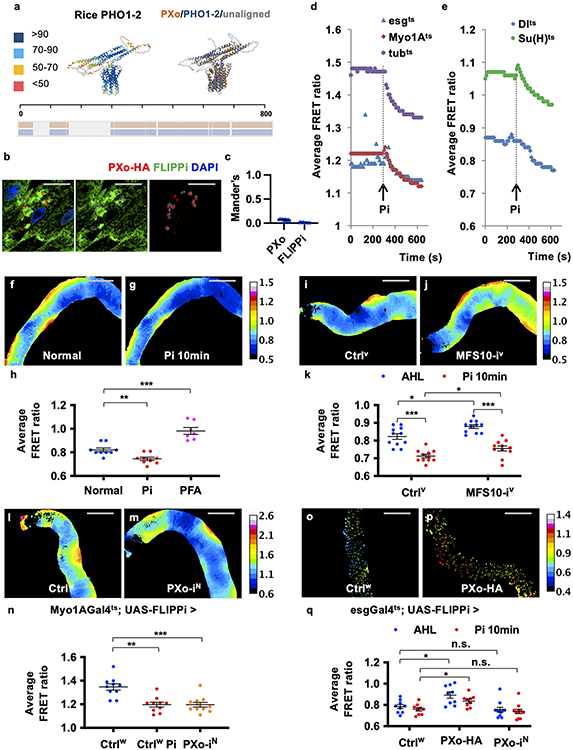
Extended Data Figure 5. Related to Figure 3. PXo structure and live imaging for cytosolic Pi.
a, AlphaFold structure prediction of rice PHO1-2, with color-scaled visualization of the prediction confidence score. The pairwise structure alignment between PXo and PHO1-2 was shown on the right, with aligned regions superposed on one another and highlighted in color. The summary of amino acid sequence alignment was shown at the bottom, with aligned regions highlighted in color. b, A single Z-stack of confocal image of midguts expressing PXo-HA and FLIPPi together in the ECs for 7d. Separate channels of HA and YFP (FLIPPi) stainings are presented on the right of merged images. c, Co-localization analysis with the quantification of the PXo-FLIPPi and FLIPPi-PXo Mander’s coefficients. N=9 per group. d, e, Kinetics of the average FRET ratio (cpVenus/CFP) in the midguts expressing FLIPPi ubiquitously (tubts) or specifically in ECs (Myo1Ats), progenitors (esgts), ISCs (Dlts) or EBs (Su(H)ts) for 3-7 days. The imaging videos were captured for ~10 min. The time points adding an extra final concentration of 30 mM Pi (~300s, using sodium phosphate or potassium phosphate) to the AHL imaging buffer is indicated by black arrows. Note that a new steady cytosolic Pi level is reached ~5 min after the addition of extra Pi. f, Color-scaled visualization of FRET ratios in the posterior midgut with ubiquitous FLIPPi expression for 7d. The same area was imaged again 10 min after the addition of 30 mM Pi (g). h, FRET ratio quantification of midguts with ubiquitous FLIPPi expression for 7d on normal food before or after 30 mM Pi addition to the imaging buffer (N=9), or with the last 4d on PFA food (N=7). P values from bottom to top: 3.91E-3, 3.50E-4. i, j, k, FRET ratio visualization and quantification of ECs expressing FLIPPi alone or FLIPPi together with MFS10 RNAi for 7d, before or 10 min after 30 mM Pi addition. N=11 per group. P values from bottom to top: 9.77E-4, 9.77E-4, 0.0173, 0.0192. l, m, n, FRET ratio visualization and quantification of midguts expressing FLIPPi alone (by crossing to Ctrlw, N=10) or FLIPPi together with PXo RNAi (NIG line, N=11) in ECs for 6d. P values from bottom to top: 1.95E-3, 4.03E-4. o, p, FRET ratios visualization of progenitors expressing FLIPPi alone or together with PXo-HA for 7d. q, FRET ratio quantification of midguts expressing FLIPPi alone (N=9), FLIPPi together with PXo-HA (N=9), or FLIPPi together with PXo RNAi (NIG line, N=11) for 7d, before or after Pi addition. P values from bottom to top: 0.0210, 0.936, 0.0114, 0.778. Data are mean ± SEM. P values are from Wilcoxon signed rank test for same sample comparison before and after Pi addition in (h, k, n), two-tailed Mann-Whitney u test between different samples (h, k, n) or one-way ANOVA with Bonferroni’s multiple comparison test (q). Scale bars, 10 μm (b), 200 μm (f, g, i, j, l, m, o, p).