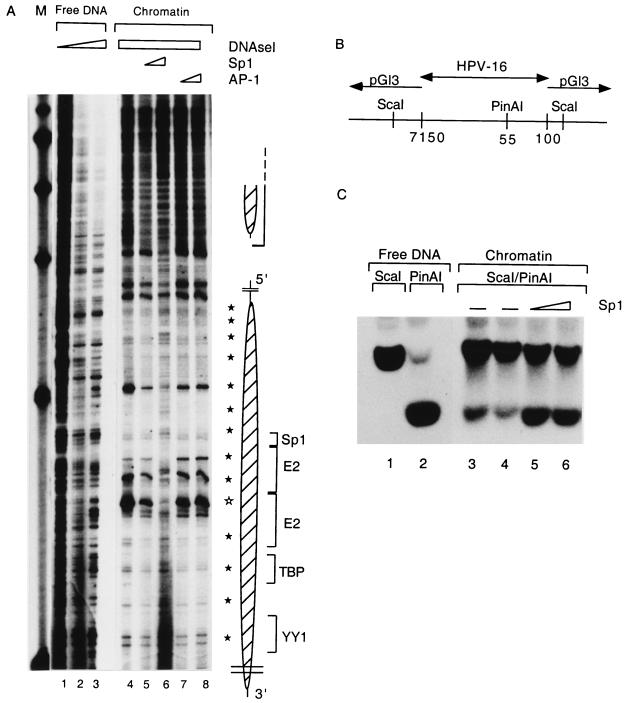
FIG. 6.
Sp1 can rearrange the structure of the E6 promoter-bound nucleosome. (A) pHPV-16-Luc DNA was assembled into chromatin, subjected to DNase I treatment, and processed as described for Fig. 3 to visualize the nucleosomal footprints overlapping with the HPV-16 E6 promoter (lanes 4 to 8) and enhancer (large and small oval shapes, respectively, on the right). Weak 10-bp-spaced bands (filled stars) indicate DNase I accessibility due to the rotational phasing of the nucleosomal core, and a strong hypersensitive site (open star) is most likely the center of the dyad symmetry of the nucleosome. As controls, lanes 1 to 3 show DNase I treatment of free HPV-16 LCR DNA. A nucleosome (Np16) protects four cis-responsive elements of the E6 promoter, namely, binding sites for Sp1, the viral factor E2, TBP, and a YY1 site (brackets on the right of the oval shape). Sp1 (lanes 5 and 6) or AP-1 (lanes 7 and 8) was added 3.5 h after start of the chromatin assembly and in the case of Sp1 resulted in the reemergence of the cleavage pattern of the free DNA and the disappearance of nucleosome-specific DNase I-hypersensitive sites. Lane M, markers. (B) Schematic representation of an HPV-16 LCR clone from position 7150 to 100 in the vector pGI3. The enzyme ScaI cleaves in vector sequences on both sides of the cloned segment; PinAI cleaves at position 55 in the promoter sequences of HPV-16. (C) Southern blot of a ScaI digest (lane 1) and a ScaI-PinAI double digest (lane 2). The ScaI-PinAI fragment is released inefficiently in the presence of chromatin (lanes 3 and 4) but efficiently with increasing Sp1 concentrations (lanes 5 and 6). This blot was processed with a radioactive probe specific for the ScaI-PinAI fragment.