Figure 3. m6A reader proteins have diverse functions and mechanisms of m6A recognition.
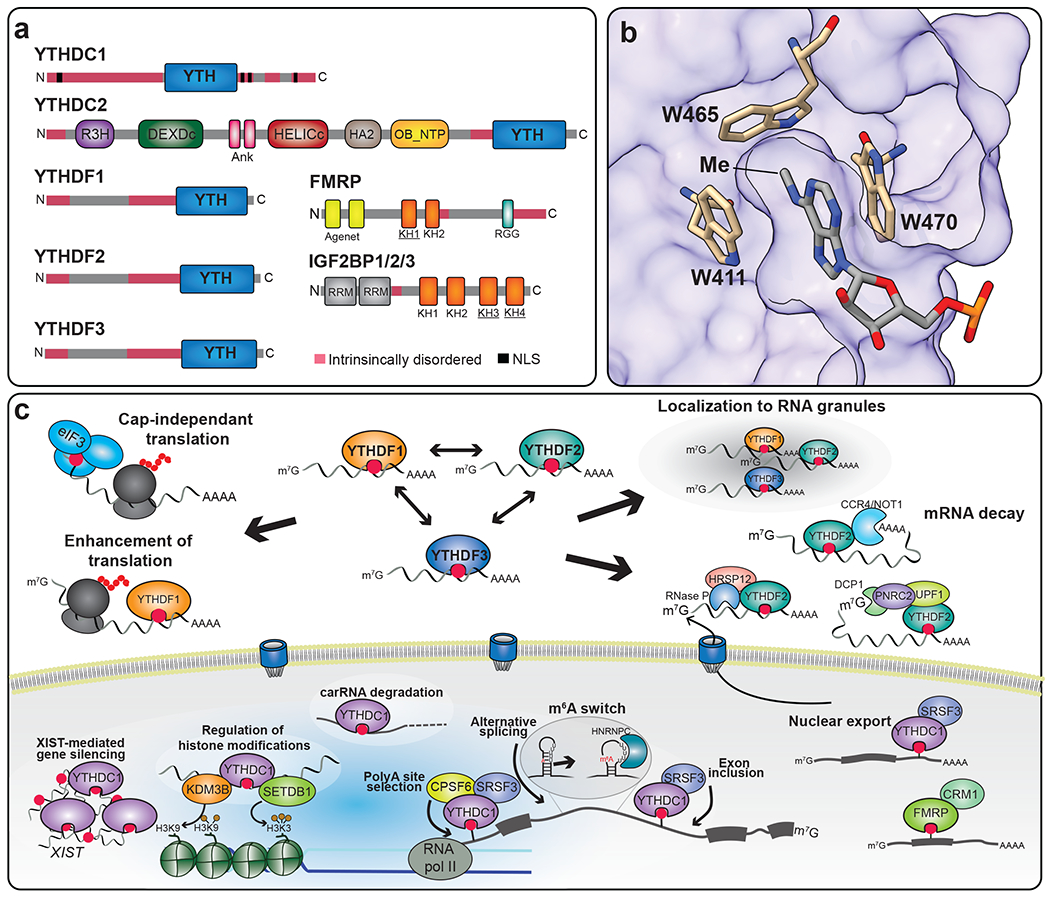
a) Schematic showing the domain structures of direct m6A binding proteins. YTHDF1/2/3, YTHDC1, and YTHDC2 recognize m6A through the highly conserved YTH domain. K-homology (KH) domains mediate m6A binding in FMRP and IGF2BP proteins. The KH domains required for m6A recognition are underlined. (b) Surface representation of the YTH domain of human YTHDF1 in complex with m6A (PDB: 4RCJ). m6A is nested in a hydrophobic pocket of the YTH domain. The three tryptophan residues forming the aromatic cage surrounding m6A are highlighted, as well as the position of the methyl moiety in m6A. c) m6A readers impact several aspects of RNA processing and function. In the nucleus, YTHDC1 regulates chromatin accessibility, promotes degradation of carRNAs, and binds to m6A sites within the lncRNA XIST to promote gene silencing. YTHDC1 also interacts with splicing factors and controls alternative splicing, polyadenylation, and nuclear export. FMRP recognition of methylated transcripts also promotes their export. In the cytoplasm, m6A is recognized by eIF3 to promote cap-independent translation of mRNAs with 5’-UTR m6A sites. YTHDF proteins can localize to stress granules and P-bodies, and all three proteins can promote mRNA decay. The role of YTHDF2 in mRNA destabilization is the most extensively studied and can include recruitment of various effector proteins to promote decapping, deadenylation, or endonucleolytic decay. Binding of YTHDF1 and YTHDF3 can also lead to increased translation of methylated mRNAs in some cell types.
