Fig. 2.
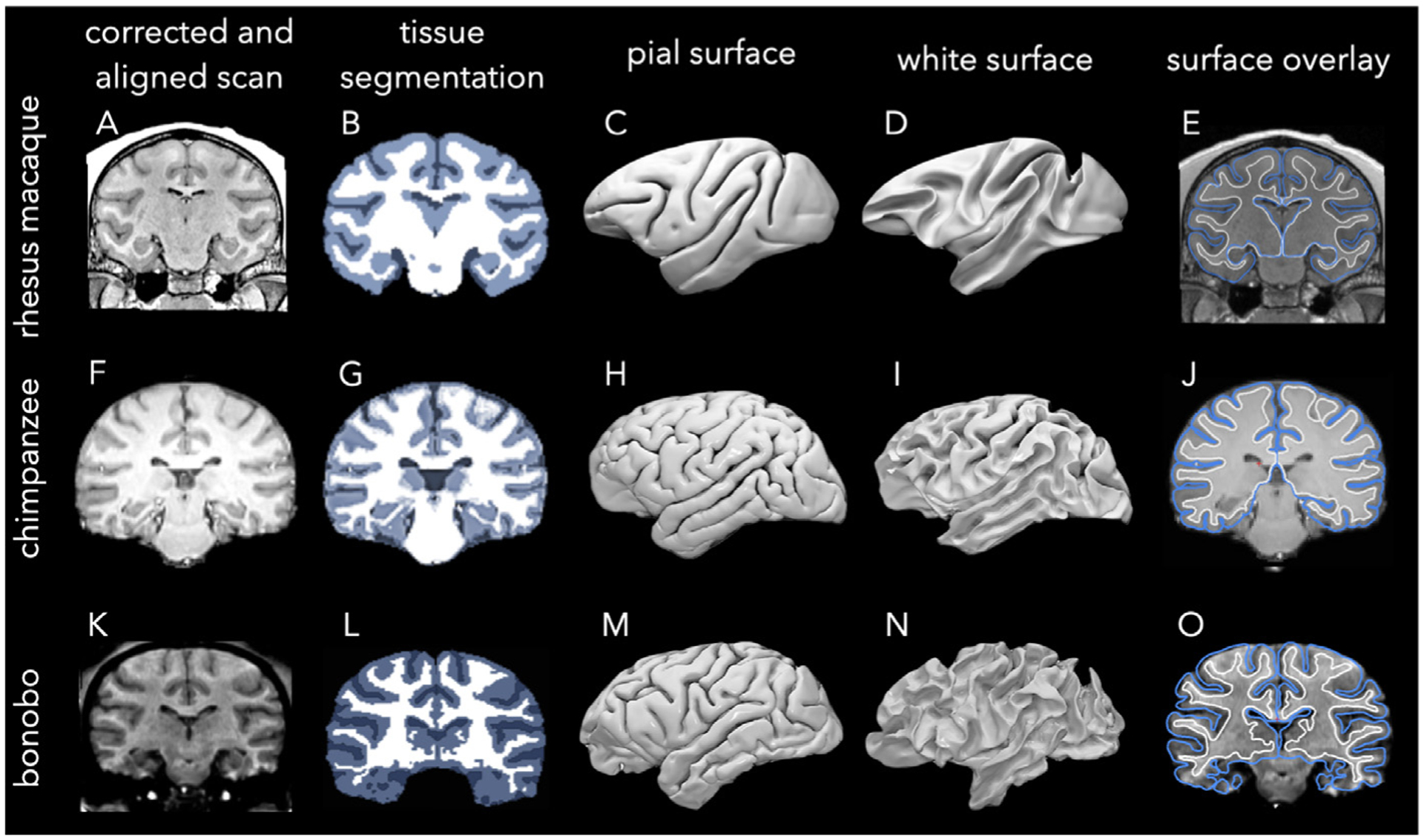
Preprocessing steps of the rhesus macaque, chimpanzee, and bonobo scans. Rhesus macaque scans from PRIME-DE were aligned to the NMT template and bias-field inhomogeneities were corrected (A) before 3-class tissue segmentation (B) and the reconstruction of pial (C) and white (D) surfaces using AFNI. Chimpanzee scans from NCBR were aligned to the Juna-Chimp template, bias-field corrected, and skull-stripped (F) before 3-class tissue segmentation (G) and reconstruction of pial (H) and white (I) surfaces using SPM and CAT. A bonobo scan from NCBR was bias-corrected, anatomically aligned, and skull-stripped (K) before 3-class tissue segmentation (L) and reconstruction of pial (M) and white (N) surfaces using BrainVisa Morphologist toolbox. The rightmost column shows the surface boundary estimations overlaid on top of MR slices for each species (E,J,O). Note that the segmentation of rhesus-macaque and chimpanzee subjects rely on the NMT and Juna-Chimp templates and species-specific segmentation methods, resulting in slight differences. For instance, the medial thalamus and lateral ventricles are within the inner compartment of the white matter surface in chimpanzees (J) but not in rhesus macaques (E) and the bonobo (O). However, this does not affect our cortical thickness estimations as thickness values that are less than 0.5 mm, including along the medial wall, are set to zero.
