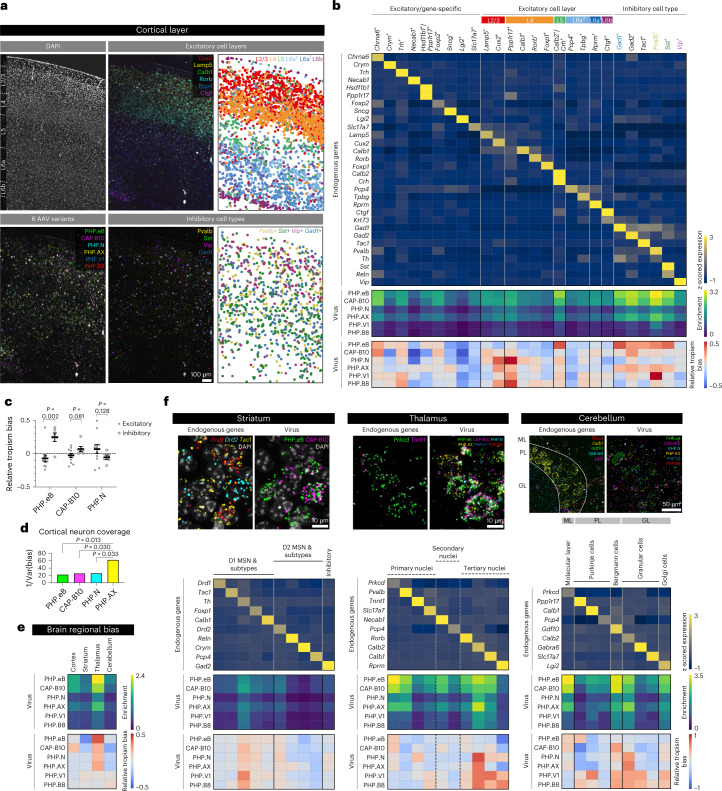
Fig. 4. Neuronal subtype tropism profiling of systemic AAVs in mouse cortical layers and other brain regions.
a, Labeling of the cortex region by DAPI and the six AAV variants. For excitatory cell layers and inhibitory subtypes, the left and middle panels show the real RNA images of selected genes acquired from the experiment, and the right panels show the cell types inferred from clustering based on endogenous gene expression. b, Endogenous (top, cividis color map) and viral gene expression profiles (enrichment: middle, viridis; relative tropism bias: bottom, coolwarm) across the cell type clusters identified. Colored cell types are visualized in a. c, The relative tropism bias of AAV-PHP.eB, AAV.CAP-B10 and AAV-PHP.N across the group of excitatory neurons (total 11 clusters) and inhibitory neurons (total six clusters; mean ± s.e.m.; two-sided unpaired t-test). d, The cortical neuron coverage of efficient variants (AAV-PHP.eB, AAV.CAP-B10, AAV-PHP.N and AAV-PHP.AX) measured by the inverse variance of relative tropism bias (the coolwarm heat map in b) across all cell type clusters (F-test on variance). We omitted AAV-PHP.V1 and AAV-PHP.B8 from this analysis as their transduction efficiency is too low to be considered for overall neuronal transduction. e, Viral expression profiles across selected mouse brain regions (cortex, striatum, thalamus and cerebellum). We separated the endogenous gene expression matrix into the fields of view of each region and used this profile to identify the regional bias of the variants. f, Endogenous and viral gene expression profiles in cell type clusters identified in the striatum, thalamus and cerebellum. We selected ten genes for striatum, ten for thalamus and nine for cerebellum that have been shown to be enriched in each region and classified cells into the clusters represented by each gene. Based on their Ward distance dendrogram, we manually merged clusters into known subtypes. Unlike the striatum and cerebellum, which are composed of genetically distinct cell types, the thalamus has relatively gradual variation in gene expression across topographical nuclei; therefore, we separated cells into three putative groups (marked by dashed lines).