Figure 2.
Blocking specific ion channels in vitro and in silico results in similar network phenotypes
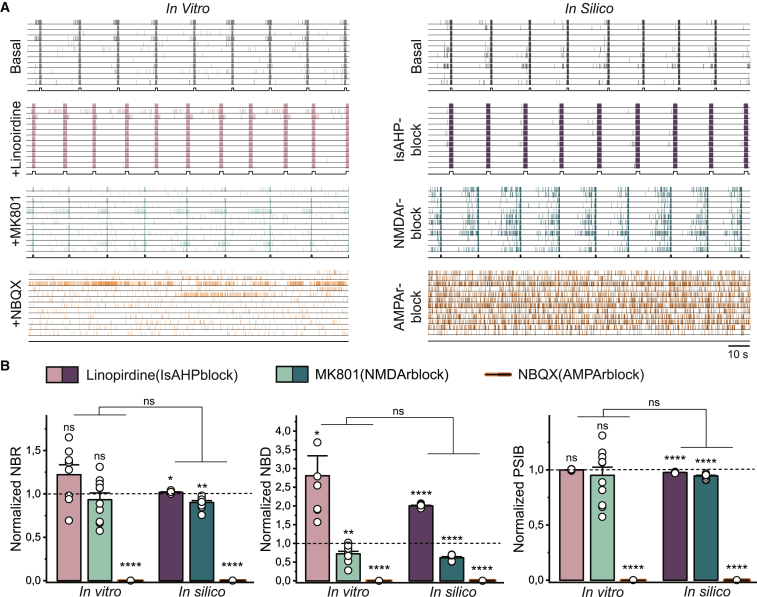
(A) Left: representative raster plots showing 100 s of spontaneous activity from a control network in vitro in basal conditions, treated with 1.5 μM linopirdine to block KCNQ potassium currents, 1 μM MK-801 to block NMDArs, and 50 μM NBQX and 10 μM NASPM to block AMPArs. Right: representative raster plots showing 100 s of simulated activity from the in silico model in basal conditions, when the conductance of the sAHP channels is halved to model the effect of linopirdine, when all the NMDArs are blocked, and when all AMPArs are blocked. Black lines at the bottom of the raster plots indicate detected NBs.
(B) Quantification of the normalized NBD, normalized NBR, and normalized PSIB in vitro and in silico with 8 networks for linopirdine (sAHPblock), 10 networks for MK-801 (NMDArblock), and 4 networks for NBQX (AMPArblock) per condition. Data represent mean ± SEM ns p > 0.05, ∗p < 0.05, ∗∗p < 0.005, ∗∗∗∗p < 0.0001. Groups were compared using a mixed effect model with multiple comparisons and Bonferroni correction.