Fig. 1. Schematic illustration of pipeline for preclinical testing of candidate therapeutic TCRs.
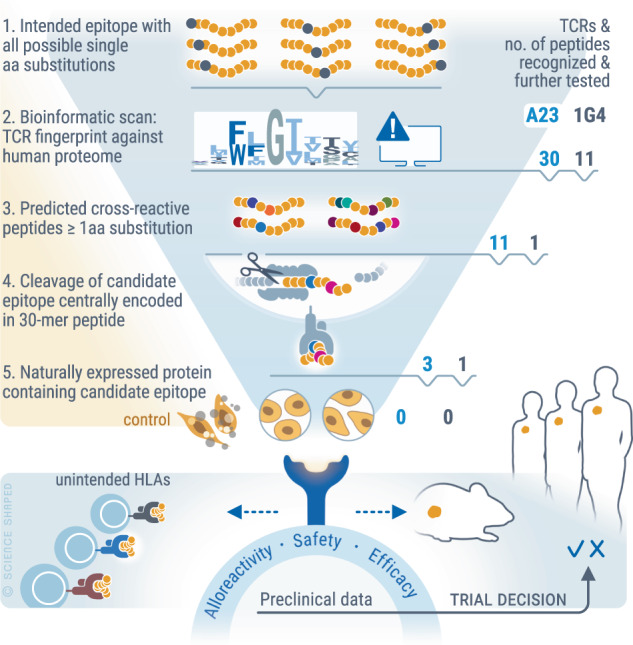
Potential off-target reactivity of candidate therapeutic TCRs is first mapped using a peptide library containing the intended epitope with all possible single AA substitutions (Step 1). The TCR fingerprint is next screened against the human proteome using a computer algorithm. This bioinformatic search identifies potential cross-reactive epitopes containing one or any combination of multiple allowed AA substitutions (Step 2), which are then synthesized and tested for TCR-recognition in Step 3. Cleavage of candidate epitopes is studied by probing TCR-reactivity against target cells electroporated with mRNA constructs encoding 30-mer peptides containing the candidate cross-recognized epitope in the middle (Step 4). To evaluate if the candidate off-target epitope is processed and presented in a physiological setting, TCR-reactivity is probed against cell lines naturally expressing confirmed high levels of proteins containing the candidate off-target epitope (Step 5). The sum of preclinical data accumulated from (i) the off-target reactivity pipeline, (ii) assessment of potential recognition of unintended HLA-alleles and (iii) in vivo efficacy studies form the basis for an informed decision regarding clinical translation. Figure 1 is created by Ellen Tenstad, Science Shaped.
