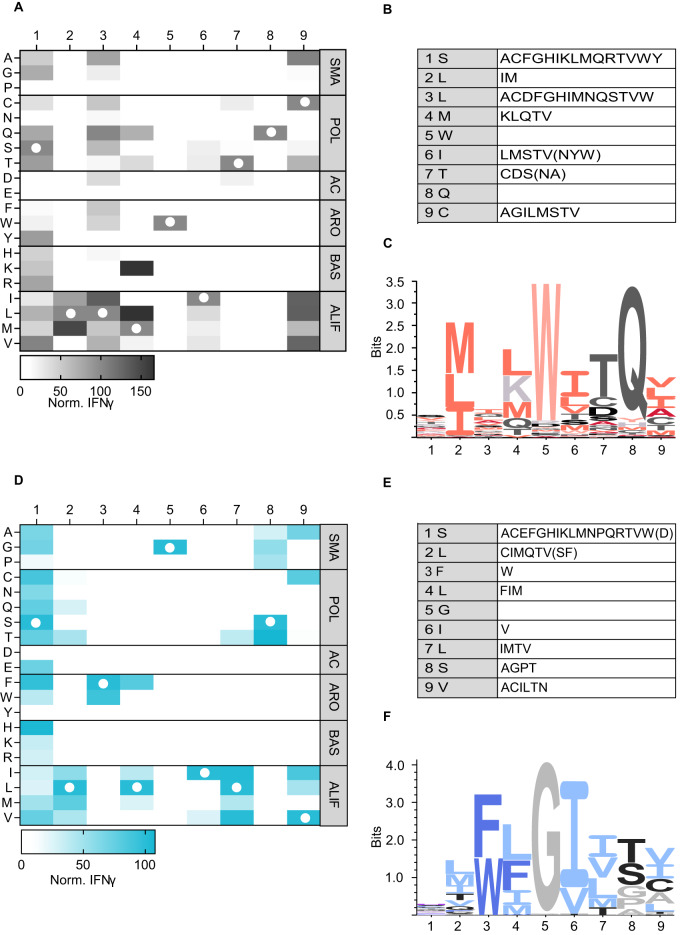
Fig. 2. TCR fingerprinting of 1G4 and A23 using a positional scanning peptide matrix and a functional readout.
1G4 TCR-engineered T cells were incubated with HLA-A2+ B-LCL cells (A), and A23 TCR-Ts were incubated with HLA-A2+ K562 cells (D) loaded with a library of 9-mer peptides containing single amino acid exchanges compared to the cognate peptide. Supernatants of 24 h co-cultures were analyzed for IFN-γ content by ELISA. IFN-γ values are normalized to the response to the original peptide. Each heatmap shows the mean of three independent experiments with one technical replicate in each. Column/row intersections indicate the replaced amino acid at a given position, and white circles show the original peptide sequence. Substitutions are divided by physicochemical properties: SMA, small; POL, polar; AC, acidic; ARO, aromatic; BAS, basic; ALIF, aliphatic. B, E Overview of amino acid exchanges recognized at different AA positions that were used to query the curated human proteome databases UniProtKB/Swiss-Prot and Protein Data Bank by the ScanProsite tool when applying a cut-off of ≥10% or ≥5% (in parenthesis) of the IFN-γ production induced by the cognate peptide. Recognition pattern of the 1G4 TCR (C) and the A23 TCR (F), visualized as a sequence logo based on the data from (A, D).