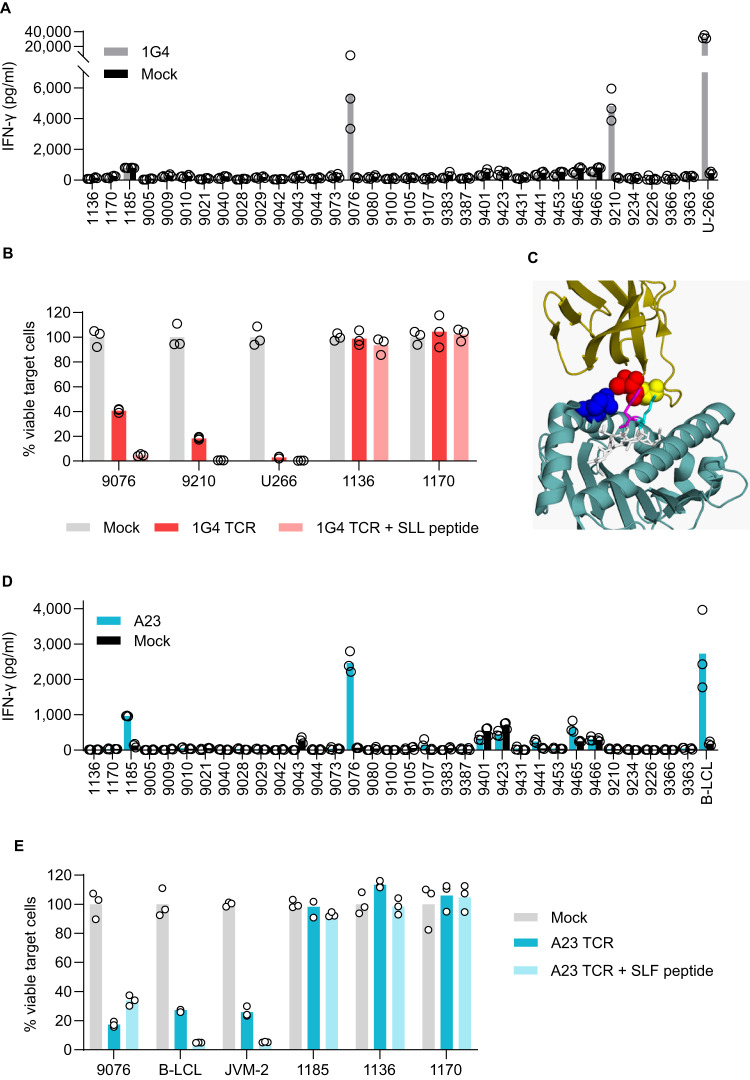
Fig. 4. Mapping the allo-reactivity profiles of 1G4 and A23 TCRs reveals cross-recognition of unintended HLA-alleles within the HLA-A02 supertype.
1G4 TCR-Ts (A) and A23 TCR-Ts (D) were co-incubated with a panel of B-LCL cells with a known HLA type (listed in Supplementary Table 5). Supernatants collected after 24 h of co-culture were analyzed for IFN-γ content by ELISA. Bars shows mean from one experiment representative of two performed with dots representing technical replicates. Positive controls: U-266 A2+ (NY-ESO-1pos) in (A) and B-LCL A2+ (CD20 pos) in (D). 1G4 TCR-Ts (B) or A23 TCR-Ts (E) were co- incubated with B-LCL lines expressing cross-reactive HLA alleles: For 1G4: 9076 (HLA-A*02:06- and HLA-A*02:07-positive) and 9210 (HLA-A*02:03-positive), for A23: 9076 (HLA-A*02:06- and HLA-A*02:07-positive) and 1185 (HLA-A*02:05-positive). After 48 h, number of surviving B-LCL cells was quantified by flow-cytometry and normalized to wells incubated with Mock cells. Loading with 100 nM of SLL or SLF peptide was included as a control. Positive controls: U266 (for 1G4) B-LCL and JVM2 (for A23). Negative controls (1136 and 1170). Figure shows one of two independent experiments performed with three technical replicates in each. Circles denote technical replicates. C Crystal structure of TCR 1G4 binding HLA-A*02:01 and NY-ESO-1157 (SLL peptide). 1G4 TCR: Ribbon model in dark yellow. Amino acids exchanged in mutant 1G4 are shown as spheres (Threonine (postion 95, red), Serine (position 96, yellow)). SLL peptide: Stick model in white. Amino acids that are in close spatial proximity to the TCRα chain of 1G4 are Methionine (position 4, cyan) and Tryptophan (position 5, magenta). HLA-A*02:01: Ribbon model in dark cyan. Conserved TCR contact residue that is in close spatial proximity to the TCRα chain of 1G4 is shown as sphere (Glutamine (position 155, blue)). Protein data bank file: 2BNR32. Visualized using PyMOL.