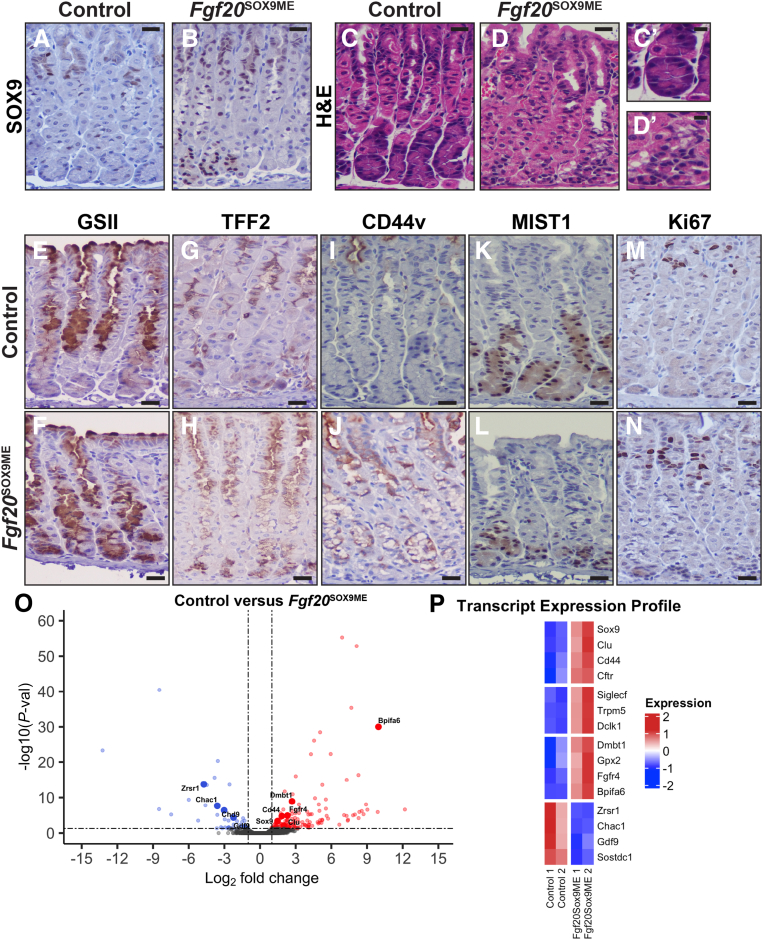
Figure 5.
Relative to postnatal misexpression, adult misexpression of Sox9 causes less-dramatic expansion of mucous cell features. (A) Anti-SOX9 shows that SOX9 is largely restricted to the neck region of adult corpus units. (B) SOX9 expression in Fgf20SOX9ME animals. In a subset of adult units, SOX9 is expressed from pit to base. (C and D) H&E shows that Fgf20SOX9ME units (D, D`) have altered base morphology with reduced cell size compared with control animals (C, C`). (E–N) Immunohistochemistry to detect representative markers of gastric lineages with hematoxylin counterstain. (E and G) Compared with control animals, Fgf20SOX9ME animals have expanded expression of mucous neck cell markers (F) GS-II lectin and (H) TFF2, extended in the base region, although less significantly compared with postnatal misexpression of Sox9. (I) Similar to postnatal tissue, CD44v was not expressed in the corpus during homeostasis. (J) However, after Sox9 misexpression, we observed marked expression of CD44v throughout the pit and neck, but not the base. (K) MIST1 strongly marks all corpus base regions in control animals. (L) After adult Sox9 misexpression, MIST1 expression is maintained in chief cells. (M and N) Adult Sox9 misexpression does not alter the proliferative center of the unit because Ki67-positive cells are still restricted to the isthmus region similar to controls. (O) Analysis of RNA-seq data from whole adult corpus of Fgf20SOX9ME animals treated with doxycycline for 2 weeks compared to control animals. Volcano plot generated from the comparison of bulk RNA-seq from adult Fgf20SOX9ME and control animals: The log2 fold change indicates the mean expression level for each gene. Blue dots, red dots, and black dots represent down-regulated genes, up-regulated genes, and insignificantly differentially expressed genes, respectively. A total of 150 genes were up-regulated, and 46 genes were down-regulated in Fgf20SOX9ME compared with controls, with the thresholds of log2 fold change greater than 1 and false discovery rate less than 0.05. (P) Heatmap (normalized transcripts per million) of 15 differentially expressed genes of interest (log2 fold change, >1; false discovery rate, <0.05) found in our RNA-seq. We detected significant up-regulation of Sox9 and a subset of metaplasia (Cd44, Clu, and Cftr) and tuft-cell–associated (Dclk1, Trpm5, and Siglecf) markers in Fgf20SOX9ME animals. Scale bars: (A–N) 20 μm, (C’ and D’) 10 μm. P-val, P value.