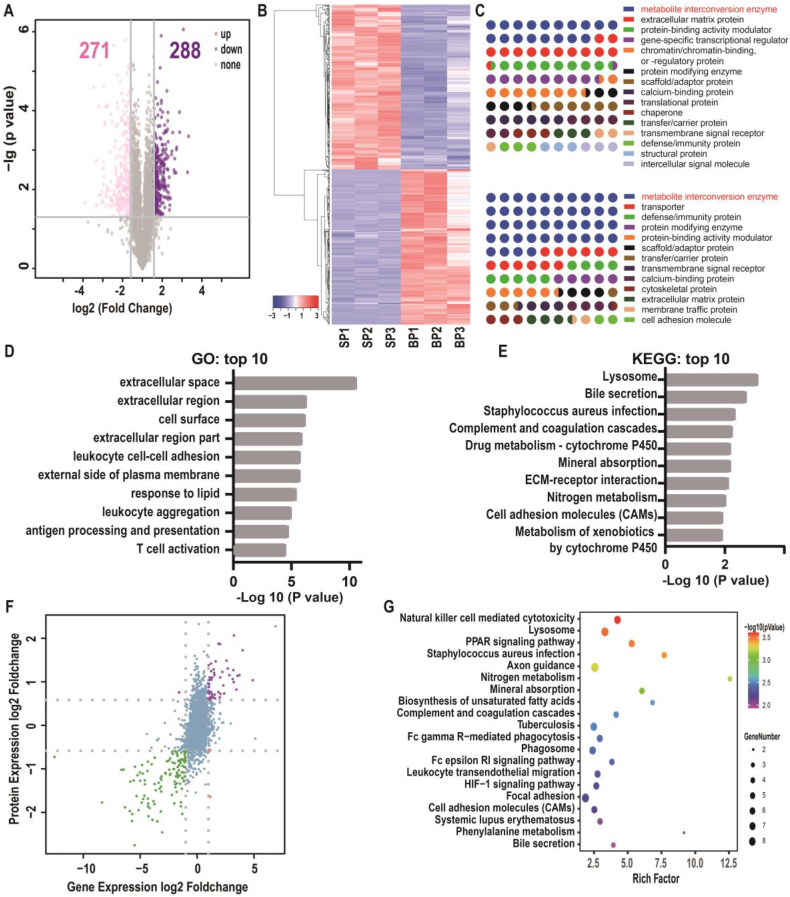
Fig. 4.
Comparative proteomics and combined transcriptomics/proteomics of SCMECs and BMECs. (A) A volcano plot of differentially expressed proteins (DEPs) in SCMECs (FC ≥ 1.5 or ≤ 0.67 for upregulated (purple) or downregulated (pink) DEPS, respectively; p-value ≤ 0.05). (B) Proteomic clustering in SCMECs (SP1–3) compared to that in BMECs (BP1–3). (C) Classification of DEPs using the PANTHER system. (D and E) Top 10 enriched GO terms (D) and KEGG pathways (E) among the DEPs. (F) A volcano plot of differentially expressed genes and proteins (DEGPs) (log2 FC > 1 or < -1 for upregulated or downregulated DEGPs, respectively) identified by combined transcriptomics/proteomics analysis. (G) A scatter plot of top 20 enriched KEGG pathways among the DEGPs.