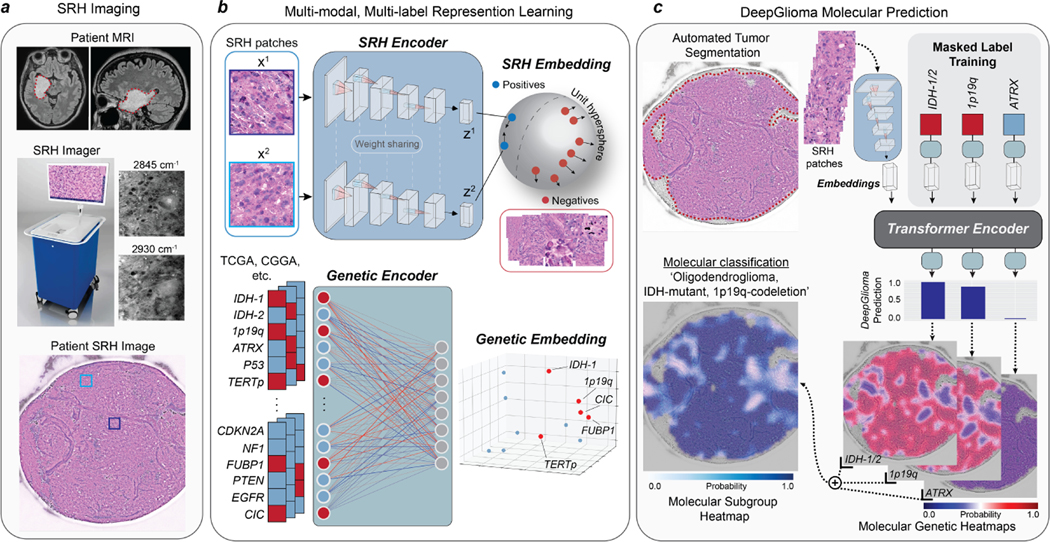
Fig. 1. Bedside SRH and DeepGlioma workflow.
a, A patient with a suspected diffuse glioma undergoes surgery for tumor biopsy or surgical resection. The SRH imaging system is portable and imaging takes place in the operating room, performed by a single technician using simple touch screen instructions. A freshly excised tissue specimen is loaded directly into a premade microscope slide and inserted into the SRH imager without the need for tissue processing (Extended Data Fig. 1). Raw SRH images are acquired at two Raman shifts, 2,845cm−1 and 2,930cm−1, as strips. The time to acquire a 3×3mm2 SRH image is approximately 90 seconds. Raw optical images can then be colored using a custom hematoxlyin and eosin (HE) virtual staining method for clinician review. b, DeepGlioma is trained using a multi-modal dataset. First, SRH images are used to train an CNN encoder using weakly supervised, multi-label contrastive learning for image feature embedding (Extended Data Fig. 3). Second, public diffuse glioma genomic data from TCGA, CGGA, and others (Extended Data Table 2) are used to train a genetic encoder to learn a genetic embedding that represents known co-occurrence relationships between genetic mutations (Extended Data Fig. 5). c, The SRH and genetic encoders are integrated into a single architecture using a transformer encoder for multi-label prediction of diffuse glioma molecular diagnostic mutations. We use masked label training to train the transformer encoder (Extended Data Fig. 6). Because our system uses patch-level predictions, spatial heatmaps can be generated for both molecular genetic and molecular subgroup predictions to improve model interpretability, identify regions of variable confidence, and associate SRH image features with DeepGlioma predictions (Extended Data Fig. 9 and 10).