Extended Data Fig 9. Molecular genetic and molecular subgroup heatmaps.
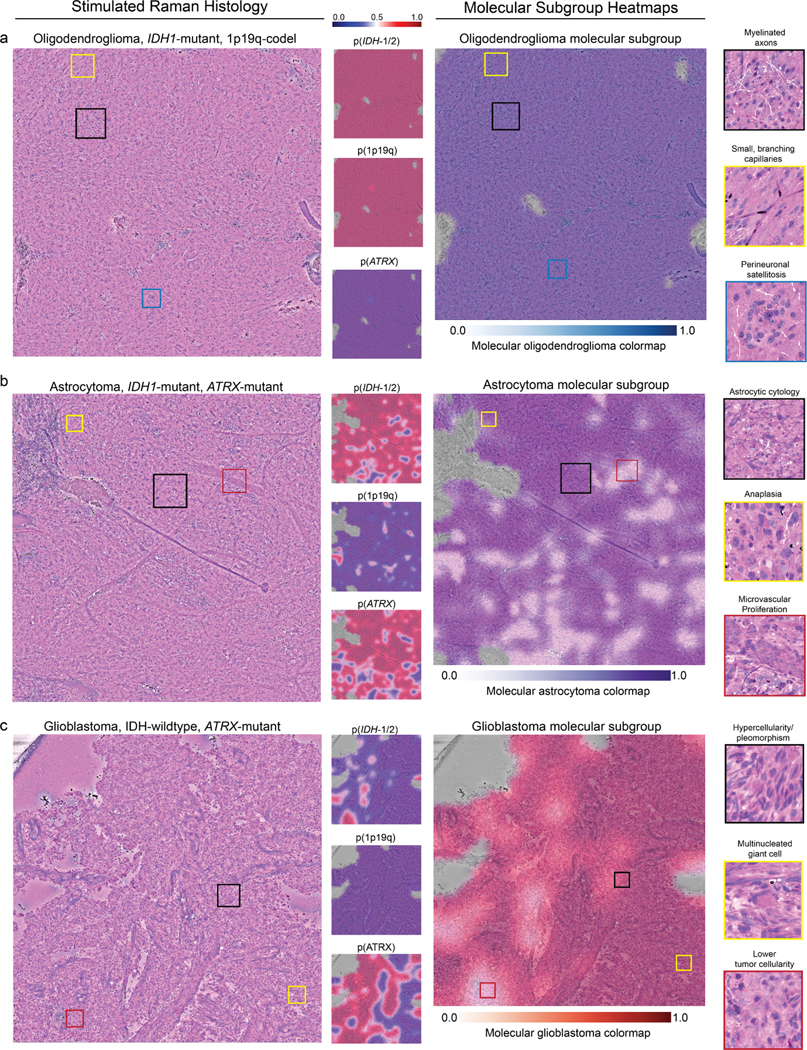
DeepGlioma predictions are presented as heatmaps from representative patients included in our prospective clinical testing dataset for each diffuse glioma molecular subgroup. a, SRH images from a patient with a molecular oligodendroglioma, IDH-mutant, 1p19q-codel. Uniform high probability prediction for both IDH and 1p19q-codel and corresponding low ATRX mutation prediction. SRH images show classic oligodendroglioma features, including small, branching ‘chicken-wire’ capillaries and perineuronal satellitosis. Oligodendroglioma molecular subgroup heatmap shows expected high prediction probablity throughout the dense tumor regions. b, A molecular astrocytoma, IDH-mutant, 1p19q-intact and ATRX-mutant is shown. Astrocytoma molecular subgroup heatmap shows some regions of lower probability that may be related to the presence of image features found in glioblastoma, such as microvascular proliferation. However, regions of dense hypercellularity and anaplasia are correctly classified as IDH mutant. These findings indicate DeepGlioma’s IDH mutational status predictions are not determined solely by conventional cytologic or histomorphologic features that correlate with lower grade versus high grade diffuse gliomas. c, A glioblastoma, IDH-wildtype tumor is shown. Glioblastoma molecular subgroup heatmap shows high confidence throughout the tumor specimen. Additionally, this tumor was also ATRX mutated, which is know to occur in IDH-wildtype tumors [10]. Despite the high co-occurence of IDH mutations with ATRX mutations, DeepGlioma was able to identify image features predictive of ATRX mutations in a molecular glioblastoma. Because ATRX mutations are not diagnostic of molecular glioblastomas, the ATRX prediction does not effect the molecular subgroup heatmap (see ‘Molecular heatmap generation‘ section in Methods). Additional SRH images and DeepGlioma prediction heatmaps can be found at our interactive web-based viewer deepglioma.mlins.org.
