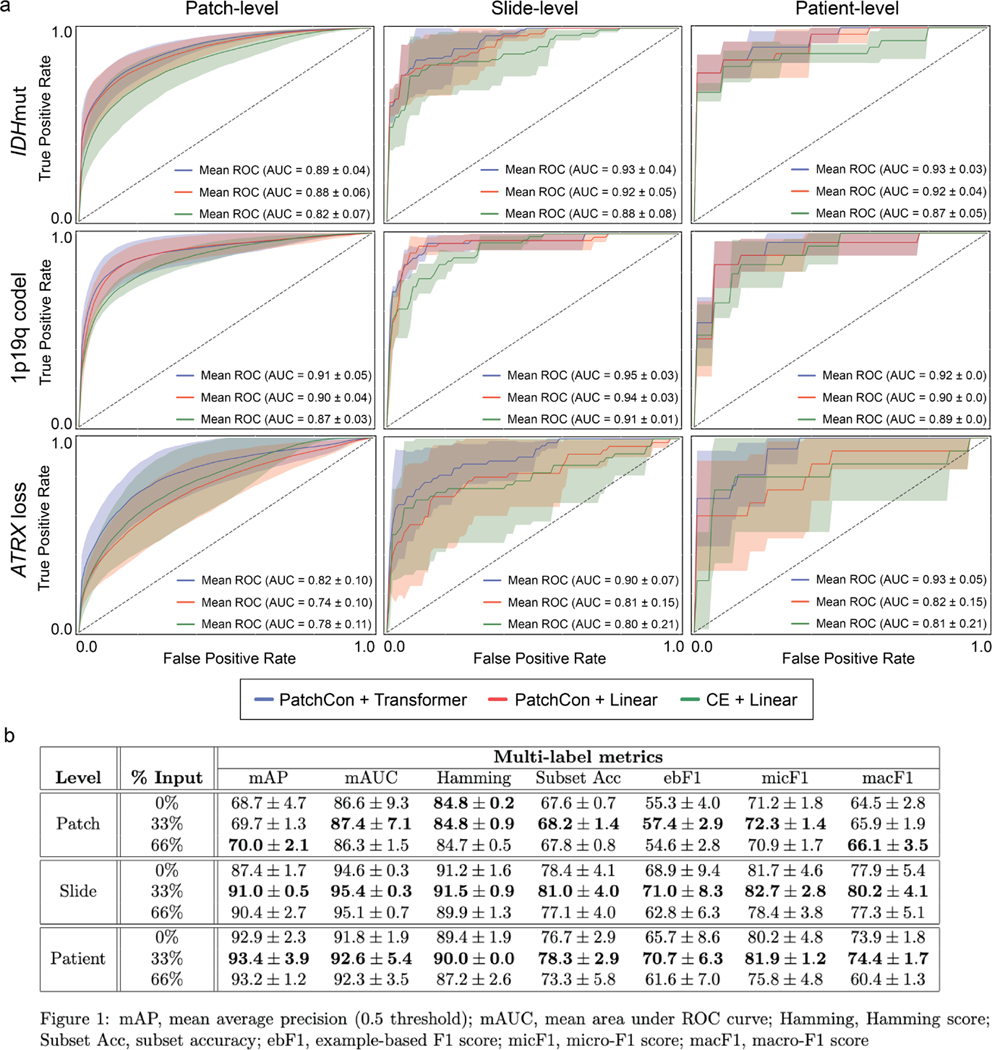
Extended Data Fig 7. Ablation studies and cross-validation results.
We conducted three main ablation studies to evaluate the following model architectural design choices and major training strategies: (1) cross-entropy versus contrastive loss for visual representation learning, (2) linear versus transformer-based multi-label classification, and (3) fully-supervised versus masked label training. a, The first two ablation studies are shown in the panel and the details of the cross-validation experiments are explained in the Methods section (see ‘Ablation Studies‘). Firstly, a ResNet50 model was trained using either cross-entropy or PatchCon. The PatchCon trained image encoder was then fixed. A linear classifier and transformer classifier were then trained using the same patchcon image encoder in order to evaluate the performance boost from using a transformer encoder. This ablation study design allows us to evaluate (1) and (2). The columns of the panel correspond to the three levels of prediction for SRH image classification: patch-, slide-, and patient-level. Each model was trained three times on randomly sampled validation sets and the average (± standard deviation) ROC curves are shown for each model. Each row corresponds to the three molecular diagnostic mutations we aimed to predict using our DeepGlioma model. The results show that PatchCon outperforms cross-entropy for visual representation learning and that the transformer classifier outperforms the linear classifier multi-label classification. Note that the boost in performance of the transformer classifier over the linear model is due to the deep multi-headed attention mechanism learning conditional dependencies between labels in the context of specific SRH image features (i.e. not improved image feature learning due to fixed encoder weights). b, We then aimed to evaluate (3). Similar to the above, a single ResNet50 model was trained using PatchCon and the encoder weights were fixed for the following ablation study to isolate the contribution of masked label training. Three training regimes were tested and are presented in the table: no masking (0%), 33% masking (one label randomly masked), and 66% (two labels randomly masked). To better investigate the importance of masked label training, we report multiple multi-label classification metrics. We found that 33% masking, or randomly masking one of three diagnostic mutations, showed the best results across all metrics at the slide-level and patient-level. We hypothesize that this results from allowing a single mutation to weakly define the genetic context while allowing supervision from the two masked labels to backpropagate through the transformer encoder.