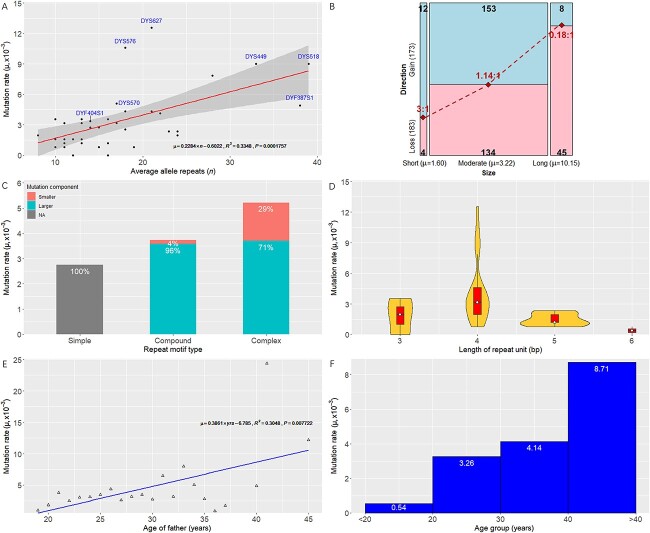
Figure 2.
Sequence-based mutation patterns at Y chromosomal short tandem repeat (Y-STRs). (A) Positive linear relationship between the mutation rate and the average allele repeat number (slope = 0.2284; R2 = 0.3348; P = 1.76 × 10−4). The initially defined rapidly mutating Y-STR markers (DYF387S1a/b, DYF404S1a/b, DYS449, DYS518, DYS570, DYS576, and DYS627) are annotated. (B) Correlation between the allele size (short, moderate, and long) and the direction of mutation (gain and loss). The average mutation rate of long alleles is significantly greater than that of moderate and short alleles (P = 2.35 × 10−12). Longer alleles present a tendency towards repeat losses whilst shorter alleles towards repeat gains. (C) Average mutation rates of different repeat motif types (simple, compound, and complex). Significant differences are observed between simple–compound (P = 0.0201), simple–complex (P = 2.35 × 10−12), and compound–complex (P = 0.0250) as well as the component of larger versus smaller units occurring mutations between compound–complex (P = 2.20 × 10−16). (D) Average mutation rates of different repeat unit lengths. A small but negative linear is observed when the repeat unit ≥4 bp (slope = −2.1243, R2 = 0.1603, P = 0.0190). (E) Positive linear relationship between the mutation rate and the age of father at the gametogenesis (slope = 0.3861; R2 = 0.3048; P = 0.0077). (F) Average mutation rates of different father’s age intervals. The number of mutations is significantly increased with the age intervals (P = 0.0048).