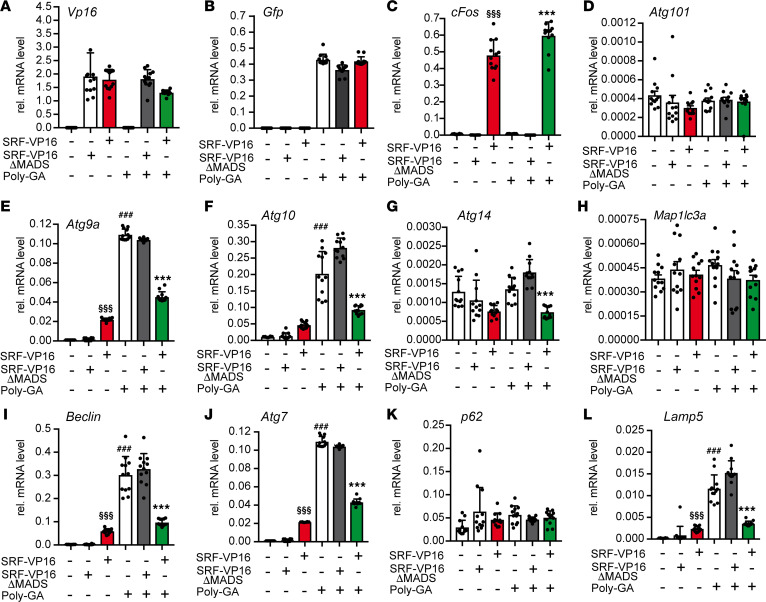
Figure 7. SRF-VP16 reduces C9orf72-associated induction of autophagy genes.
HEK293 cells expressed either constitutively active SRF-VP16 or inactive SRF-VP16ΔMADS in the presence or absence of aggregates formed by Poly-GA expression. Subsequently, qPCR was performed to assess mRNA abundance of the genes indicated. (A–C) Expression of Vp16 tagged SRF (A) and Gfp tagged Poly-GA (B) was similar on an mRNA level. SRF-VP16 but not SRF-VP16ΔMADS induced cFos (C). (D–L) SRF-VP16 but not SRF-VP16ΔMADS induced Atg9a (E), Atg10 (F), Beclin (I), and Atg7 (J) as well as Lamp5 (L). Poly-GA expression upregulated Atg9a (E), Atg10 (F), Beclin (I), Atg7 (J), and Lamp5 (L) but not Atg101 (D), Atg14 (G), Map1lc3a (H), and p61 (K). SRF-VP16 but not SRF-VP16ΔMADS downregulated several of those autophagy- and lysosome-encoding genes induced by Poly-GA aggregate formation. In A–L, n values are indicated by each black dot reflecting 1 cell culture dish. *, #, § denote significance between SRF-VP16 and SRF-VP16ΔMADS in the presence of Poly-GA (fifth and sixth bar), between mock and Poly-GA (first and fourth bar), and between SRF-VP16 and SRF-VP16ΔMADS (second and third bar), respectively. *P < 0.05, **P < 0.01, ***P < 0.001. Statistical testing was performed by 1-way ANOVA with Tukey corrections.