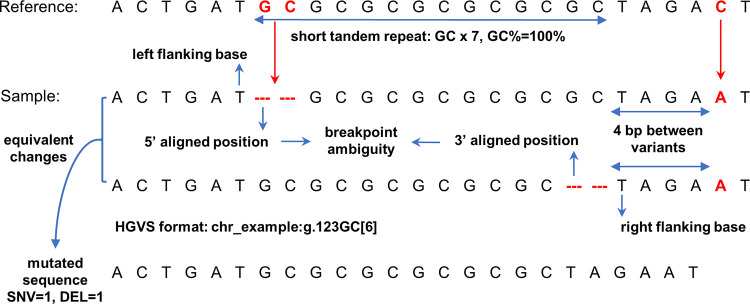
Fig 1. The illustration of sequence contexts of a genomic variant.
The example shows the reference and sample sequences where a dinucleotide GC is deleted at “chr_example”, position 123. The deletion is located in a short tandem repeat region, which is a dinucleotide repeat motif GC with a copy number of seven. The GC content (GC%) of the short tandem repeat region is 100%. The short tandem repeat can result in the deletion having multiple possible representations but all lead to an equivalent change. This issue is also known as the breakpoint ambiguity, which indicates the exact breakpoint of a variant is impossible to be confidently identified. The Human Genome Variation Society (HGVS) recommends describing different types of variants with specific roles and formats. The left and right flanking bases of the deletion are marked based on these equivalent deletions on the sequence. There is also a single nucleotide substitution located in the 3’ direction of this deletion. The distance between two variants is 4 bp, which is also determined from on these equivalent deletions. The mutated sequence can be determined by considering all the variants within the region, which in the above example are one single nucleotide variant (SNV) and one deletion (DEL). The variant annotations related to the sequence contexts are shown in bold text.