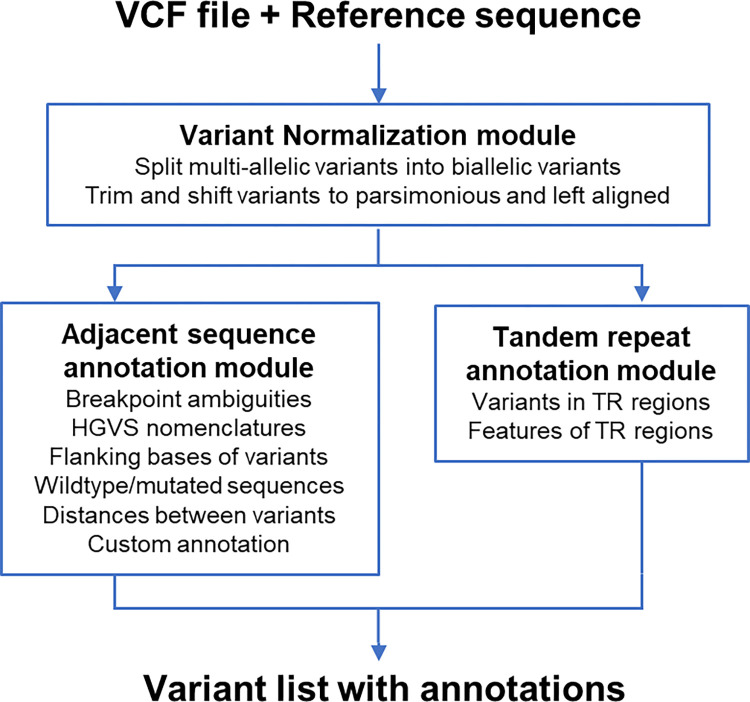
Fig 2. VarSCAT workflow.
The input VCF file is first passed to the variant normalization module of VarSCAT, from which the essential information, including positions, reference/alternative alleles, identifiers, and genotypes of variants are extracted. This module can split any potential multiallelic variants into biallelic variants and then normalizes all input variants as parsimonious and left aligned. The output of the variant normalization module is passed to an adjacent sequence annotation module and a tandem repeat (TR) annotation module. The adjacent sequence annotation module can be used to annotate the breakpoint ambiguities, flanking bases of variants, wildtype/mutated DNA sequences, HGVS nomenclature, distances between adjacent variants, and custom annotations. The tandem repeat annotation module can annotate tandem repeat regions of input variants.