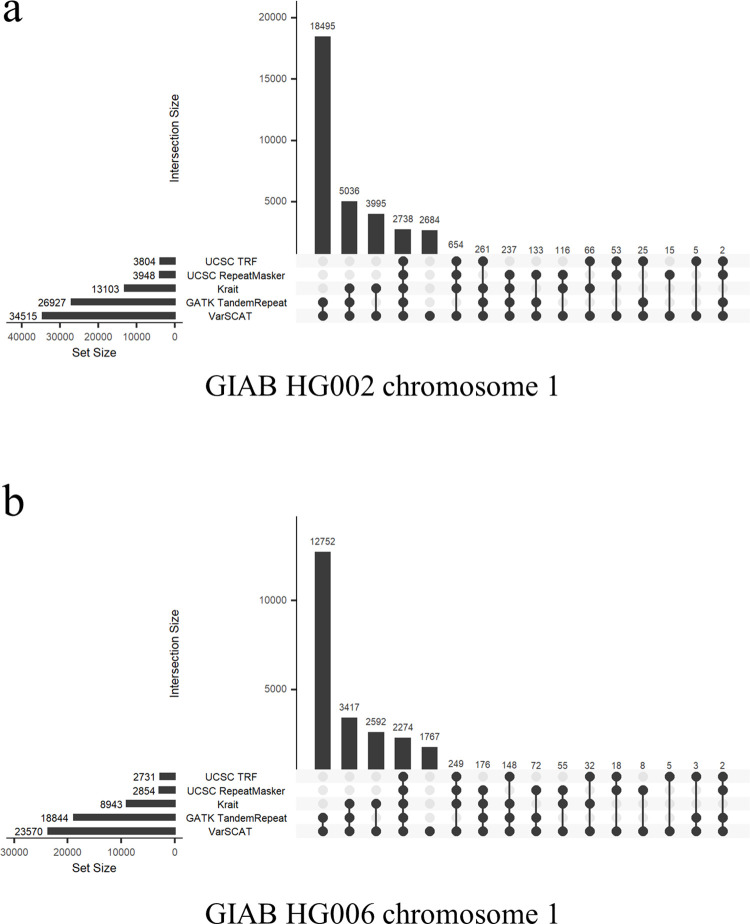
Fig 3. The benchmarking results of the VarSCAT tandem repeat annotation module.
Benchmarking was performed for annotating small variants in perfect STR regions in chromosome 1 of (a) GIAB HG002, and (b) GIAB HG006. GATK ‘TandemRepeat’ function is an annotation method that directly takes a VCF file as the input; TRF and RepeatMasker from the UCSC Genome Browser’s ‘Simple Repeats’ and ‘RepeatMasker’ tracks represent a ready-made STR annotation approach; Krait is an annotation method for detecting perfect STRs with a reference genome.