ABSTRACT
Sequencing is increasingly used for infective endocarditis (IE) diagnosis. Here, the performance of 16S rRNA gene PCR/sequencing of heart valves utilized in routine clinical practice was compared with conventional IE diagnostics. Subjects whose heart valves were sent to the clinical microbiology laboratory for 16S rRNA gene PCR/sequencing from August 2020 through February 2022 were studied. A PCR assay targeting V1 to V3 regions of the 16S rRNA gene was performed, followed by Sanger and/or next-generation sequencing (NGS) (using an Illumina MiSeq), or reported as negative, depending on an algorithm that included the PCR cycle threshold value. Fifty-four subjects, including 40 with IE, three with cured IE, and 11 with noninfective valvular disease, were studied. Thirty-one positive results, 11 from NGS and 20 from Sanger sequencing, were generated from analysis of 16S rRNA gene sequence(s). Positivity rates of blood cultures and 16S rRNA gene PCR/sequencing of valves were 55% and 75%, respectively (P = 0.06). In those with prior antibiotic exposure, positivity rates of blood cultures and 16S rRNA gene PCR/sequencing of valves were 11% and 76%, respectively (P < 0.001). Overall, 61% of blood culture-negative IE subjects had positive valve 16S rRNA gene PCR/sequencing results. 16S rRNA gene-based PCR/sequencing of heart valves is a useful diagnostic tool for pathogen identification in patients with blood culture-negative IE undergoing valve surgery in routine clinical practice.
KEYWORDS: 16S rRNA gene, infective endocarditis, Sanger sequencing, next-generation sequencing
INTRODUCTION
Infective endocarditis (IE) is associated with significant morbidity and mortality (1, 2). Identification of causative microorganisms guides optimal antimicrobial agent selection, treatment duration, and decisions to operate and the associated surgical approach (2). While blood cultures may define the microbiological etiology of IE, up to 40% of endocarditis cases have negative blood cultures (2, 3). Blood culture-negative endocarditis (BCNE) may occur due to nonviable microorganisms (e.g., as a result of administration of antimicrobial agents), or fastidious microorganisms that do not grow well in blood cultures (3–5). Multimodal diagnostic approaches, including serological and/or molecular methods, may be useful in BCNE (4, 5).
PCR targeting the 16S rRNA gene has been used to detect microorganisms causing IE in excised valve tissue (3, 4, 6–10). Shrestha et al. retrospectively evaluated excised fresh valves from 174 subjects undergoing surgery for active IE, who had definite IE based on modified Duke criteria, using 16S and/or 28S rRNA PCR followed by Sanger sequencing (8). Valve sequencing was more likely to be positive than valve culture in those with IE (90% versus 31%, P < 0.001) and yielded fewer positive results than valve culture in those without IE (3% versus 33%, P < 0.001) (8). Fournier et al. evaluated 16S and/or 18S rRNA PCR assays followed by Sanger sequencing on valve tissue, identifying a microorganism in 63% (476/759) of BCNE cases (4). Sanger sequencing has limitations, including the inability to resolve mixed sequences and low sensitivity when microorganisms are present in low abundance (11). Next-generation sequencing (NGS) provides sequence data for large numbers of individual DNA molecules in parallel and is being increasingly used in infectious disease diagnostics; recent reports highlight the potential utility of NGS-based approaches for IE diagnosis (12–15). There are two NGS-based approaches, targeted metagenomic sequencing (tMGS), focusing on specific genes, such as the 16S rRNA gene, and shotgun metagenomic sequencing (sMGS), which instead sequences all input nucleic acid. Santibanez et al. evaluated tMGS targeting of the 16S rRNA gene V3-V4 region on valve tissues from 27 patients with IE compared with conventional methods (blood cultures and/or valve PCR) (14). tMGS showed concordance with conventional methods in 89% (24/27), identifying one potential new pathogen and detecting potentially mixed infections. tMGS using the 16S rRNA gene is unable to detect fungi, viruses, and parasites; in contrast, unbiased sMGS can theoretically detect any microorganism. Cheng et al. (15) evaluated sMGS on valve tissue from 51 subjects undergoing heart surgery (41 with definite IE), reporting a sensitivity and specificity of 98% and 86%, respectively; valve sMGS was more sensitive than culture-based methods (49% versus 98%, P < 0.0001) and detected fastidious or unculturable microorganisms (six Coxiella burnetii, one Bartonella quintana, and one Haemophilus parainfluenzae isolate). Only bacteria were identified by sMGS in this study, suggesting that tMGS could have been performed similarly (15). However, sMGS detected mecA in one case of Staphylococcus aureus infection, a finding not provided by tMGS. In contrast to tMGS, sMGS requires deeper sequencing. Further, the complexity of presequencing workflows and overall sequencing costs, alongside bioinformatics and interpretive needs, are greater for sMGS than tMGS (11, 16).
In August 2020, a PCR assay targeting the 16S rRNA gene, with amplicon sequencing using Sanger or NGS approaches, or reported as negative, depending upon an algorithm using cycle threshold (Ct) values, was introduced into the routine clinical practice at Mayo Clinic. An analysis of 2,146 sterile tissues or body fluids clinically tested on Mayo Clinic and Mayo Clinic Laboratories patients using this approach showed higher positivity than culture (53% versus 42%, P < 0.001) (17). Here, the diagnostic performance of this approach was evaluated on fresh and formalin-fixed paraffin-embedded (FFPE) valve tissues of Mayo Clinic patients being evaluated for IE, with molecular results compared to conventional culture.
MATERIALS AND METHODS
Study design.
This retrospective study was performed at Mayo Clinic (Rochester, MN). The study population included Mayo Clinic patients aged ≥18 years whose heart valves (fresh or FFPE) were clinically tested by 16S rRNA gene PCR followed by Sanger and/or NGS from August 2020 through February 2022. The study was approved by the Mayo Clinic Institutional Review Board (no. 20-012373) with a waiver of consent. Clinical information was obtained by review of the electronic medical record and included subject demographics, underlying disease, and clinical manifestations. A subset of the subjects was included in reference (17).
Definitions.
Subjects were classified as having IE, cured IE, or noninfective valvular disease (NIVD) based on the 2023 Duke-ISCVID criteria (2). Those who had completed endocarditis treatment within 6 months of testing with no antibiotics administered for at least 6 weeks before surgery were categorized as cured IE. Time between antibiotic initiation and diagnostic testing was determined by calculating days between initiation of an antibiotic active against the cause of IE and valve resection or blood culture collection. If more than one valve sample from the same subject was tested, only one sample was analyzed, prioritizing the positive sample. Valve tissue cultures were considered positive if there was any growth. Histopathological evidence of endocarditis included evidence of active or healing endocarditis.
Sample processing.
Blood and tissue cultures were performed following standards of care. 16S rRNA gene PCR/Sanger sequencing and/or NGS was performed as described previously (17). Briefly, DNA was extracted on an EMAG system (bioMérieux) and amplified on a LightCycler 480II (Roche Diagnostics) using dual priming oligonucleotides targeting the 16S rRNA V1 to V3 regions. Amplified DNA was sequenced by Sanger sequencing and/or NGS (Illumina MiSeq), or reported as negative, depending on an algorithm that incorporated PCR crossing threshold (Ct) value and melting temperature, as described previously (17). Demultiplexed results were analyzed using Pathogenomix bioinformatics software and interpreted by a team of expert molecular bacteriology technologists overseen by board-certified doctoral-level clinical microbiologists with expertise in molecular bacteriology.
Statistical analysis.
Sensitivity and specificity were calculated using 2023 Duke-ISCVID criteria (2) as the gold standard to define or rule out IE. Data were analyzed using MedCalc statistical software (version 18.11.3). Results were expressed as means ± standard deviations (SDs) or as medians (interquartile ranges [IQRs]). Comparisons were performed using chi-square or Fisher’s exact tests. Sensitivity and specificity of blood and tissue culture and those of 16S rRNA gene PCR/sequencing were compared using McNemar’s tests.
RESULTS
Overall, 55 cases were considered for inclusion, of which one was excluded as it was not classifiable. Among the 54 included subjects, 40 (74%) had IE, three (6%) had cured IE, and the remaining 11 (20%) had NIVD. Demographic and clinical characteristics are summarized in Table 1. Among the 40 IE subjects, 39 (98%) had definite IE and one (3%) had possible IE.
TABLE 1.
Demographic and clinical characteristics of infection and noninfected patients
| Characteristic | Value for group |
||
|---|---|---|---|
| Noninfective valvular disease, n = 11 | Infective endocarditis, n = 40 | Cured infective endocarditis,a n = 3 | |
| Age, mean yr ± SD | 63.5 ± 10.8 | 60.0 ± 14.0 | 62.0 ± 5.3 |
| Sex, male, no. (%) | 6 (54.5) | 25 (62.5) | 2 (66.7) |
| Caucasian, no. (%) | 9 (81.8) | 35 (87.5) | 2 (66.7) |
| Native valve, no. (%) | 6 (54.5) | 13 (32.5) | 3 (100) |
| Prosthetic valve, no. (%) | 5 (45.5) | 27 (67.5) | 0 |
| Body mass index, kg/m2, median (IQR) | 26.7 (20.6–32.5) | 27.4 (23.3–34.0) | 32.0 (22.1–40.2) |
| Predisposing heart conditions, no. (%) | |||
| Valvular disease | 7 (63.6) | 16 (40.0) | 3 (100) |
| Structural | 5 (45.5) | 7 (17.5) | 3 (100) |
| Congenital | 1 (9.1) | 6 (15.0) | 0 |
| Other | 1 (9.1) | 3 (7.5) | 0 |
| Prosthetic vascular graft | 2 (18.2) | 17 (42.5) | 0 |
| Previous infective endocarditis | 1 (9.1) | 7 (17.5) | 3 (100) |
| Associated medical conditions, no. (%) | |||
| Congestive heart failure | 5 (45.5) | 10 (25.0) | 1 (33.3) |
| Diabetes mellitus | 1 (9.1) | 5 (12.5) | 1 (33.3) |
| Chronic immunosuppressive therapy | 1 (9.1) | 4 (10.0) | 0 |
| Active neoplastic disease | 0 | 3 (7.5) | 0 |
| Hemodialysis dependence | 1 (9.1) | 2 (5.0) | 1 (33.3) |
| Injection drug use | 0 | 2 (5.0) | 0 |
| Antibiotics administered at time of blood culture collection, no. (%) | 1 (9.1) | 9 (22.5) | 0 |
| Antibiotics administered within 4 wk prior to surgery, no. (%) | 1 (9.1) | 33 (82.5) | 1 (33.3) |
| Valvular involvement, no. (%) | |||
| Aortic valve only | 7 (63.6) | 25 (62.5) | 1 (33.3) |
| Mitral valve only | 2 (18.2) | 5 (12.5) | 2 (66.7) |
| Tricuspid valve only | 1 (9.1) | 2 (5.0) | 0 |
| Pulmonary valve only | 0 | 0 | 0 |
| Multiple valves | 0 | 8 (20.0)b | 0 |
Cases with cured infective endocarditis include patients diagnosed with infective endocarditis within 6 months who had completed antibiotic treatment, including subjects with Streptococcus anginosus, Streptococcus mutans, and Staphylococcus aureus isolated from blood culture (one case each).
Six cases involving the aortic and mitral valves and two cases involving the mitral and tricuspid valves.
Heart valve 16S rRNA gene PCR/sequencing.
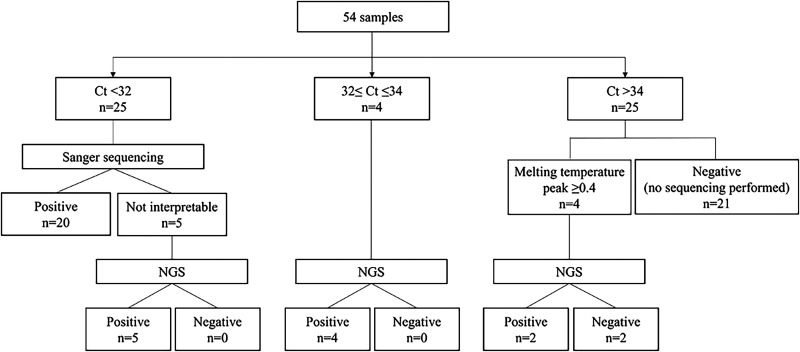
Twenty-five (46%) samples had Ct values of <32 cycles and underwent Sanger sequencing, per the algorithm, with 20 (80%) yielding reportable, positive results (Fig. 1). The other five yielded uninterpretable results with Sanger sequencing and, along with four samples with Ct values of ≥32 and ≤34 cycles (7%, 4/54 of all samples), were sent to NGS, all yielding positive results. Among the 25 samples (46%) with Ct values of >34 cycles, four with a melting temperature peak of ≥0.4 underwent NGS, of which two were reported as positive (Fig. 1). Overall, 31 valves were positive by 16S rRNA gene PCR/sequencing, 20 by Sanger sequencing, and 11 by NGS. The addition of NGS to Sanger sequencing resulted in positivity of 57% (31/54) compared with 37% (20/54) with Sanger sequencing alone, although the latter was not performed on six NGS-positive samples with Ct values of ≥32 cycles.
FIG 1.
Sample flowchart. Ct, cycle threshold; NGS, next-generation sequencing.
All IE cases were monomicrobial; microorganisms detected by 16S rRNA gene PCR/sequencing are summarized in Table S1 in the supplemental material. Thirty IE subjects (of 40; 75%) had positive 16S rRNA gene PCR/sequencing results, as did one subject (of 3; 33%) with cured IE and none with NIVD. Sensitivities of 16S rRNA gene PCR/sequencing and blood cultures were 75% and 55%, respectively (P = 0.06) (Table 2). Specificities of the two methods were 100% and 91%, respectively (P = 0.31).
TABLE 2.
Comparison of diagnostic performances of microbiologic tests
| Method | No. positive |
Sensitivity (%) (95% CIa) | Specificity (%) (95% CI) | ||
|---|---|---|---|---|---|
| Infective endocarditis (n = 40) | Noninfective valvular disease (n = 11) | ||||
| Culture | |||||
| Valve | 13 | 0 | 33 (19–49) | 100 (72–100) | |
| Blood | 22 | 1 | 55 (38–71) | 91 (59–100) | |
| Valve 16S rRNA gene PCR/sequencing | 30 | 0 | 75 (59–86) | 100 (72–100) | |
| Fresh tissue | 26 of 35 | 0 of 9 | 74 (57–87) | 100 (66–100) | |
| Formalin-fixed paraffin-embedded tissue | 4 of 5 | 0 of 2 | 80 (28–99) | 100 (16–100) | |
| Valve Gram stain | |||||
| Fresh tissue (performed in microbiology laboratory) | 5 of 35 | 0 of 8 | 14 (5–30) | 100 (63–100) | |
| Formalin-fixed paraffin-embedded tissue (performed in pathology laboratory) | 9 of 35 | 0 of 9 | 26 (12–43) | 100 (66–100) | |
| Valve histopathology | |||||
| Active endocarditis | 22 of 34 | 0 of 9 | 65 (46–80) | 100 (67–100) | |
CI, confidence interval.
Among the 40 IE cases, 35 cases had 16S rRNA gene PCR/sequencing performed on fresh tissue and the other five had it performed on FFPE tissue. Sensitivity of 16S rRNA gene PCR/sequencing on the two specimen types was 74% and 80%, respectively (P = 0.78).
Heart valve 16S rRNA gene PCR/sequencing compared to blood cultures.
Among the 40 IE cases, microorganisms were detected in 22 (55%) by blood cultures and 30 (75%) by 16S rRNA gene PCR/sequencing; results (positive or negative) were concordant between the two methods in 25 (63%) (Table 3). In the IE group, 24 with Ct values of <32 and four with Ct values of ≥32 and ≤34 were positive by 16S rRNA gene PCR/sequencing; two with Ct values of >34 were also positive.
TABLE 3.
Performance of heart valve 16S rRNA gene PCR/sequencing versus blood culture
| Case classification | No. of samples | No. (%) |
||
|---|---|---|---|---|
| Identical findings | Organisms not detected by 16S rRNA gene PCR/sequencing | New organisms detected by 16S rRNA gene PCR/sequencing | ||
| Noninfective valvular disease | 11 | 10 (91) | 1 (9)a | 0 |
| Infective endocarditis | 40 | 25 (63) | 4 (10) | 11 (28) |
| Cured infective endocarditis | 3 | 1 (33) | 2 (67) | 0 |
Staphylococcus aureus was detected in one of two blood cultures in a single case clinically diagnosed with uncomplicated S. aureus bacteremia in which S. aureus was not considered a cause of endocarditis.
Of 22 blood culture-positive IE cases, microorganisms were detected by 16S rRNA gene PCR/sequencing in 19 (86%) (Table 4). Except in one case, microorganisms detected by the two methods were identical. One patient receiving antibiotic suppression for 2 years for Staphylococcus epidermidis endocarditis last detected by blood culture 7 months prior underwent repeat aortic root replacement with aortic valve replacement due to a worsening pseudoaneurysm; Pseudomonas aeruginosa was detected by 16S rRNA gene PCR/sequencing and tissue culture. The most common microorganism identified by both methods was Streptococcus species. In four cases (two with S. aureus and one each with S. epidermidis and Corynebacterium amycolatum), bacteria were detected by blood cultures but not 16S rRNA gene PCR/sequencing (Table 5). All four cases were diagnosed with IE prior to surgery and had received antibiotics for more than 3 months before surgery.
TABLE 4.
Characteristics of infective endocarditis subjects according to blood culture results
| Characteristic | Total no. | No. by blood culture result: |
|
|---|---|---|---|
| Positive (n = 22) | Negative (n = 18) | ||
| 16S rRNA gene PCR/sequencing | |||
| Positive | 30 | 19a | 11 |
| Negative | 10 | 3 | 7 |
| Antibiotic treatment prior to blood culture | 9 | 1 | 8 |
| Known microbiology | 35 | 22 | 13 |
| Streptococcus species | 10 | 8 | 2 |
| Streptococcus salivarius group | 3 | 3 | 0 |
| Streptococcus sanguinis | 3 | 2 | 1 |
| Streptococcus mitis group | 2 | 1 | 1 |
| Streptococcus agalactiae | 1 | 1 | 0 |
| Streptococcus mutans | 1 | 1 | 0 |
| Staphylococcus species | 8 | 7 | 1 |
| Staphylococcus epidermidis | 4 | 3a | 1 |
| Staphylococcus aureus complex | 3 | 3 | 0 |
| Staphylococcus lugdunensis | 1 | 1 | 0 |
| Enterococcus faecalis | 2 | 2 | 0 |
| Cutibacterium acnes | 5 | 0 | 5 |
| Bartonella species | 3 | 0 | 3 |
| Other | 7 | 5b | 2c |
| Unknown microbiology | 5 | 0 | 5 |
Results of the two tests were identical except in one case in which Staphylococcus epidermidis was found in blood culture and Pseudomonas aeruginosa was detected by 16S rRNA gene PCR/sequencing and valve tissue culture.
One case each involving Cardiobacterium hominis, Corynebacterium amycolatum, Granulicatella adiacens, Mycobacterium avium complex, and Parvimonas micra.
One case each involving Haemophilus parainfluenzae and Histoplasma capsulatum.
TABLE 5.
Discrepant results between heart valve 16S rRNA gene PCR/sequencing and blood culture
| Discrepancy between methods | New or missed identifications by heart valve 16S rRNA gene PCR/sequencing vs blood culture (no.) |
|---|---|
| Endocarditis organism detected by blood culture but not 16S rRNA gene PCR/sequencing | Staphylococcus aureus (2), Corynebacterium amycolatum (1), Staphylococcus epidermidis (1) |
| Endocarditis organism detected by 16S rRNA gene PCR/sequencing but not blood culture | Cutibacterium acnes (4), Bartonella species (3), Haemophilus parainfluenzae (1), S. epidermidis (1), Streptococcus mitis group (1), Streptococcus sanguinis (1) |
| Microorganism not detected by 16S rRNA gene PCR/sequencing in treated infective endocarditis | Streptococcus anginosus (1), Streptococcus mutans (1) |
| Microorganism detected by 16S rRNA gene PCR/sequencing in treated infective endocarditis | S. aureus complex (1) |
Of the 18 BCNE cases, eight (44%) were receiving antibiotics at the time of blood collection (Table 4). Eleven potential pathogens (61%) were detected by 16S rRNA gene PCR/sequencing (four by Sanger sequencing and seven by NGS), including Cutibacterium acnes (4/11, 36%) and Bartonella species (3/11, 27%) and single detections of H. parainfluenzae, S. epidermidis, Streptococcus mitis group, and Streptococcus sanguinis. Of these new identifications, three (two C. acnes and one S. epidermidis) had the same organism identified by valve culture and three (with Bartonella species detected) had IgG titers against Bartonella henselae of ≥1:800. Among the seven BCNE cases with negative 16S rRNA gene PCR/sequencing results, microorganisms were found by other diagnostic methods in two cases. One case had a positive Histoplasma immunodiffusion result and consistent findings by histopathology. The other had valve culture positive for C. acnes. No pathogen was found in the other five subjects, which included one receiving antibiotic suppression for 16 years for enterococcal endocarditis, one receiving oral antibiotic suppression for 1 year for Streptococcus mitis periprosthetic joint infection, one with S. aureus peritonitis 8 months prior to IE diagnosis with treatment completed 6 months prior to IE diagnosis, one with Tetralogy of Fallot with a bioprosthetic pulmonary valve placed 20 years prior to IE diagnosis and biliary sepsis treated with cholecystectomy 3 months prior to IE diagnosis, and one with an underlying monoclonal gammopathy, and a shotgun metagenomic sequencing test on plasma positive for Streptococcus mutans in the context of active native valve endocarditis, with Gram-positive cocci on Gram stain of the heart valve.
Heart valve 16S rRNA gene PCR/sequencing compared to blood cultures in cured IE.
Three cases with IE who had completed antibiotic treatment within 6 months and stopped antibiotics at least 6 weeks prior to valve surgery had had Streptococcus anginosus, S. mutans, and S. aureus isolated from blood cultures in one case each. One had S. aureus complex identified by heart valve 16S rRNA gene PCR/sequencing (Table 3).
Noninfective valvular disease.
16S rRNA gene PCR/sequencing was negative in all 11 NIVD cases; all had Ct values of >34. One of these subjects had S. aureus isolated from one of two blood cultures initially but not in blood cultures conducted over six consecutive days (Table 3). This was clinically considered an uncomplicated bacteremia and not S. aureus endocarditis.
Heart valve 16S rRNA gene PCR/sequencing compared to heart valve culture.
Thirteen subjects with IE had positive valve cultures (Table 2); there were no positive valve cultures in the NIVD group. Sensitivities of 16S rRNA gene PCR/sequencing and valve cultures were 75% and 33%, respectively (P < 0.001) (Table 2). 16S rRNA gene PCR/sequencing resulted in identical results (positive or negative) in 19 of 40 (48%) IE cases, with additional detections in 20 cases (see Table S2 in the supplemental material). One case with C. acnes identified by valve culture, but not by 16S rRNA gene PCR/sequencing, was associated with an indolent clinical course. The subject underwent repeat aortic root replacement with no findings of infection prior to surgery and had an infected pseudoaneurysm found intraoperatively.
Heart valve 16S rRNA gene PCR/sequencing compared to histopathological examination.
Compared with histopathologic findings of endocarditis, sensitivity of heart valve 16S rRNA gene PCR/sequencing was similar (75% versus 65%, P = 0.33) (Table S3).
Heart valve 16S rRNA gene PCR/sequencing compared to valve Gram stain.
In IE cases in which Gram staining was performed, the positivity rate of Gram staining performed in the clinical microbiology laboratory was 14% (5/35), lower than that of 16S rRNA gene PCR/sequencing (77%, 27/35; P < 0.001), and the positivity of histopathology Gram staining was 26% (9/35), also lower than that of 16S rRNA gene PCR/sequencing (74%, 26/35; P < 0.001).
Antecedent antimicrobial exposure.
Mean length of antimicrobial treatment in subjects with IE before surgery was 18 days (IQR, 6 to 38 days). 16S rRNA gene PCR/sequencing was positive in 76% (25/33) of IE subjects with prior antibiotic exposure, including 33% (6/18) with BCNE. One of nine IE subjects who received antecedent antibiotics had positive blood cultures; 16S rRNA gene PCR/sequencing of valves was less affected by prior antibiotic administration than were blood cultures (P < 0.001).
DISCUSSION
Rapid advances in sequencing-based diagnostics are improving the diagnostic yield in IE. 16S rRNA gene PCR/sequencing, including NGS, provides a tool for defining potential causative microorganisms for IE (11–14). Because of the cost and technical complexity of NGS, a tMGS-based approach was implemented clinically using a combination of PCR/Sanger sequencing, PCR/NGS, or just PCR depending on Ct value and melting temperature (16). Here, the diagnostic value of this approach, performed on heart valves, was compared to those of blood and valve culture, valve Gram stain, and valve histopathology for IE detection. 16S rRNA gene PCR/sequencing had higher sensitivity than blood cultures in the subset of subjects with prior antibiotic exposure (P < 0.001) and was a useful diagnostic tool for pathogen identification in BCNE.
Few studies have explored the diagnostic value of NGS in IE diagnosis, and published studies have included different patient populations and sequencing methods than those reported here (12, 13, 15). Zeng et al. compared sMGS of valves to blood and valve culture in 110 patients undergoing valve surgery (99 with IE) and showed sMGS to be more sensitive (86%) than blood or tissue culture (29% and 16%, respectively), but specificity was low (73%) (13). Only a small proportion of reads generated from sMGS typically correspond to potential pathogens, especially when performed on tissue which has substantial human background reads (11). Cell-free DNA by sMGS from plasma has been shown to identify causative pathogens in 87% of 23 patients with definite IE (18). tMGS of valve tissue may be preferred over sMGS when the cost and complexity (including interpretive complexity) of methods are considered, acknowledging that nonbacterial causes of IE would be missed by tMGS targeting only the 16S rRNA gene. We previously reported results of tMGS on 395 sonicate fluid samples (208 with periprosthetic joint infection) generated from resected hip or knee prosthesis, showing this to be a potential diagnostic tool for culture-negative cases as well as polymicrobial infections, with performance characteristics similar to those of sMGS (16). Like periprosthetic joint infection, IE is primarily bacterial, so tMGS may be sufficient.
As NGS is culture independent, it should be able to detect difficult-to-culture microorganisms. In the present study, 16S rRNA gene PCR/sequencing detected a microorganism in 61% of BCNE cases. In the IE group receiving antimicrobial therapy prior to surgery, 16S rRNA gene PCR/sequencing was more sensitive than valve culture (76% versus 27%, P < 0.001). Halavaara et al. showed that more than 2 weeks of preoperative antimicrobial therapy reduced valve bacterial PCR yield (53% versus 91%; P < 0.001) (19). In the current study, in those treated with antibiotics for more than 2 weeks, 16S rRNA gene PCR/sequencing positivity decreased compared to positivity in those treated for less than 2 weeks, although the difference was not statistically significant, possibly due to the sample size (14/21, 67%, versus 11/12, 92%; P = 0.11). In a prior study, tMGS was more sensitive than culture in the group that received antimicrobial therapy in the 4 weeks before sampling (63% versus 41%; P < 0.001), and adding NGS to Sanger sequencing increased positivity by 87% compared with Sanger sequencing alone for testing normally sterile tissues and body fluids (98% for tissues, 66% for fluids) (17). In the present study, heart valve NGS increased positivity by 55% compared with Sanger sequencing alone, although six specimens positive by NGS were not tested with Sanger sequencing (as a result of their having high Ct values) (20); whether such heart valve samples might have yielded usable Sanger sequencing results is unknown.
FFPE can preserve cellular morphology for years. However, nucleic acid degradation during fixation can theoretically affect the quality and quantity of DNA extracted from FFPE tissue. Compared with fresh tissue, however, despite the lack of statistical significance, and small specimen numbers, the positivity rate of FFPE tissue was similar in the current study (P = 0.78), possibly due to the abundance of microorganisms present in valves affected by IE.
There was one case in which Gram-positive cocci and active endocarditis were present by histopathology but no microorganism was detected by any diagnostic method, including 16S rRNA gene PCR/sequencing. The subject had been diagnosed with culture-negative IE and treated with antibiotics 3 weeks before surgery.
Compared to valve 16S rRNA gene PCR/sequencing, sensitivity of valve culture was low (75% versus 33%, respectively, P = 0.0001). Shrestha et al. reported higher false-positive rates with valve culture than sequencing (33% versus 3%) (8). In the present study, there were two cases in which different microorganisms were found between 16S rRNA gene PCR/sequencing and valve culture; in one case, C. acnes was isolated as a single colony in valve culture while Bartonella species was detected by 16S rRNA gene PCR/sequencing with concomitant serologic evidence of B. henselae infection. In the other case, Staphylococcus hominis and S. epidermidis were isolated from broth medium by valve culture, whereas C. acnes was detected by 16S rRNA gene PCR/sequencing. This subject had been treated with vancomycin and ceftriaxone for culture-negative prosthetic valve endocarditis for 6 weeks prior to surgery.
There were five cases in which C. acnes was identified. C. acnes has been reported to cause 4 to 8% of prosthetic valve endocarditis (21, 22). Cutibacterium species require prolonged incubation, including for blood cultures (21). C. acnes endocarditis is associated with males with underlying prosthetic, most commonly aortic, valves. In the present study, C. acnes was detected by 16S rRNA gene PCR/sequencing (n = 4) and/or valve culture (n = 3), but not blood culture; however, blood cultures were incubated for only 5 days. All cases were males with prosthetic valves, four involving aortic valves, and all had indolent clinical courses.
There are several limitations to this study, including its retrospective design and low sample number. Not all patients undergoing valve surgery had 16S rRNA gene PCR/sequencing performed, introducing selection bias; it is likely that molecular testing was preferentially ordered in BCNE. Our institution is a tertiary care referral center, introducing referral bias. Another limitation is that the test used detects only bacteria. Fungi are rare endocarditis causes, typically occurring in those with risks such as malignancy, injection drug use, or having prosthetic valves (3). A previous study showed fungal sequencing to offer no additional value beyond information provided by histopathology and blood cultures (8). In this study, however, there was one Histoplasma endocarditis case, diagnosed by serology and histopathology, and it was missed by 16S rRNA gene PCR/sequencing. It should be noted that sensitivity and specificity were calculated based on microbial detection or lack thereof and not on detection of the known cause of IE. Finally, it should be noted that sequencing was performed and sequencing type was chosen based on an algorithm depending on the Ct and melting temperature and that a team of bacteriology sequencing experts interpreted results. Findings may not generalize to all laboratories. Our experience to date is that some Sanger sequencing results can be protocolized for reporting by molecular bacteriology technologists, without review by board-certified doctoral-level clinical microbiologists with expertise in molecular bacteriology but that this is not easily done for NGS. Hopefully, improvements in analytics and understanding of performance over time will alleviate this situation.
In conclusion, 16S rRNA gene PCR/sequencing of heart valves should be prioritized in the clinical algorithm of testing for BCNE with negative Bartonella species and C. burnetii serologies.
Footnotes
Supplemental material is available online only.
Contributor Information
Robin Patel, Email: patel.robin@mayo.edu.
Patricia J. Simner, Johns Hopkins University
REFERENCES
- 1.Thuny F, Grisoli D, Collart F, Habib G, Raoult D. 2012. Management of infective endocarditis: Challenges and perspectives. Lancet 379:965–975. doi: 10.1016/S0140-6736(11)60755-1. [DOI] [PubMed] [Google Scholar]
- 2.Fowler VG, Durack DT, Selton-Suty C, Athan E, Bayer AS, Chamis AL, Dahl A, DiBernardo L, Durante-Mangoni E, Duval X, Fortes C, Fosbol E, Hannan MM, Hasse B, Hoen B, Karchmer AW, Mestres CA, Petti CA, Pizzi MN, Preston SD, Roque A, Vandenesch F, van der Meer JTM, van der Vaart TW, Miro JM. 2023. The 2023 Duke-ISCVID criteria for infective endocarditis: Updating the modified Duke criteria. Clin Infect Dis doi: 10.1093/cid/ciad271. [DOI] [Google Scholar]
- 3.Liesman RM, Pritt BS, Maleszewski JJ, Patel R. 2017. Laboratory diagnosis of infective endocarditis. J Clin Microbiol 55:2599–2608. doi: 10.1128/JCM.00635-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fournier PE, Thuny F, Richet H, Lepidi H, Casalta JP, Arzouni JP, Maurin M, Celard M, Mainardi JL, Caus T, Collart F, Habib G, Raoult D. 2010. Comprehensive diagnostic strategy for blood culture-negative endocarditis: A prospective study of 819 new cases. Clin Infect Dis 51:131–140. doi: 10.1086/653675. [DOI] [PubMed] [Google Scholar]
- 5.Houpikian P, Raoult D. 2005. Blood culture-negative endocarditis in a reference center: etiologic diagnosis of 348 cases. Medicine (Baltimore) 84:162–173. doi: 10.1097/01.md.0000165658.82869.17. [DOI] [PubMed] [Google Scholar]
- 6.Breitkopf C, Hammel D, Scheld HH, Peters G, Becker K. 2005. Impact of a molecular approach to improve the microbiological diagnosis of infective heart valve endocarditis. Circulation 111:1415–1421. doi: 10.1161/01.CIR.0000158481.07569.8D. [DOI] [PubMed] [Google Scholar]
- 7.Goldenberger D, Kunzli A, Vogt P, Zbinden R, Altwegg M. 1997. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J Clin Microbiol 35:2733–2739. doi: 10.1128/jcm.35.11.2733-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shrestha NK, Ledtke CS, Wang H, Fraser TG, Rehm SJ, Hussain ST, Pettersson GB, Blackstone EH, Gordon SM. 2015. Heart valve culture and sequencing to identify the infective endocarditis pathogen in surgically treated patients. Ann Thorac Surg 99:33–37. doi: 10.1016/j.athoracsur.2014.07.028. [DOI] [PubMed] [Google Scholar]
- 9.Marin M, Munoz P, Sanchez M, Del Rosal M, Alcala L, Rodriguez-Creixems M, Bouza E, Group for the Management of Infective Endocarditis of the Gregorio Marañón Hospital . 2007. Molecular diagnosis of infective endocarditis by real-time broad-range polymerase chain reaction (PCR) and sequencing directly from heart valve tissue. Medicine (Baltimore) 86:195–202. doi: 10.1097/MD.0b013e31811f44ec. [DOI] [PubMed] [Google Scholar]
- 10.Pettersson GB, Hussain ST. 2019. Current AATS guidelines on surgical treatment of infective endocarditis. Ann Cardiothorac Surg 8:630–644. doi: 10.21037/acs.2019.10.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiu CY, Miller SA. 2019. Clinical metagenomics. Nat Rev Genet 20:341–355. doi: 10.1038/s41576-019-0113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai S, Yang Y, Pan J, Miao Q, Jin W, Ma Y, Zhou C, Gao X, Wang C, Hu B. 2021. The clinical value of valve metagenomic next-generation sequencing when applied to the etiological diagnosis of infective endocarditis. Ann Transl Med 9:1490. doi: 10.21037/atm-21-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng X, Wu J, Li X, Xiong W, Tang L, Li X, Zhuang J, Yu R, Chen J, Jian X, Lei L. 2022. Application of metagenomic next-generation sequencing in the etiological diagnosis of infective endocarditis during the perioperative period of cardiac surgery: A prospective cohort study. Front Cardiovasc Med 9:811492. doi: 10.3389/fcvm.2022.811492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santibanez P, Garcia-Garcia C, Portillo A, Santibanez S, Garcia-Alvarez L, de Toro M, Oteo JA. 2021. What does 16S rRNA gene-targeted next generation sequencing contribute to the study of infective endocarditis in heart-valve tissue? Pathogens 11:34. doi: 10.3390/pathogens11010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng J, Hu H, Fang W, Shi D, Liang C, Sun Y, Gao G, Wang H, Zhang Q, Wang L, Wu H, Hu L, Chen L, Zhang J, Lee S, Wang F, Zhou Z. 2019. Detection of pathogens from resected heart valves of patients with infective endocarditis by next-generation sequencing. Int J Infect Dis 83:148–153. doi: 10.1016/j.ijid.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Hong HL, Flurin L, Thoendel MJ, Wolf MJ, Abdel MP, Greenwood-Quaintance KE, Patel R. 2023. Targeted versus shotgun metagenomic sequencing-based detection of microorganisms in sonicate fluid for periprosthetic joint infection diagnosis. Clin Infect Dis 76:e1456–e1462. doi: 10.1093/cid/ciac646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flurin L, Wolf MJ, Mutchler MM, Daniels ML, Wengenack NL, Patel R. 2022. Targeted metagenomic sequencing-based approach applied to 2146 tissue and body fluid samples in routine clinical practice. Clin Infect Dis 75:1800–1808. doi: 10.1093/cid/ciac247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eichenberger EM, Degner N, Scott ER, Ruffin F, Franzone J, Sharma-Kuinkel B, Shah P, Hong D, Dalai SC, Blair L, Hollemon D, Chang E, Ho C, Wanda L, de Vries CR, Fowler VG, Ahmed AA. 2023. Microbial cell-free DNA identifies the causative pathogen in infective endocarditis and remains detectable longer than conventional blood culture in patients with prior antibiotic therapy. Clin Infect Dis 76:e1492–e1500. doi: 10.1093/cid/ciac426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halavaara M, Martelius T, Jarvinen A, Antikainen J, Kuusela P, Salminen US, Anttila VJ. 2019. Impact of pre-operative antimicrobial treatment on microbiological findings from endocardial specimens in infective endocarditis. Eur J Clin Microbiol Infect Dis 38:497–503. doi: 10.1007/s10096-018-03451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fida M, Khalil S, Abu Saleh O, Challener DW, Sohail MR, Yang JN, Pritt BS, Schuetz AN, Patel R. 2021. Diagnostic value of 16S ribosomal RNA gene polymerase chain reaction/Sanger sequencing in clinical practice. Clin Infect Dis 73:961–968. doi: 10.1093/cid/ciab167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fida M, Dylla BL, Sohail MR, Pritt BS, Schuetz AN, Patel R. 2019. Role of prolonged blood culture incubation in infective endocarditis diagnosis. Eur J Clin Microbiol Infect Dis 38:197–198. doi: 10.1007/s10096-018-3397-1. [DOI] [PubMed] [Google Scholar]
- 22.Lindell F, Soderquist B, Sundman K, Olaison L, Kallman J. 2018. Prosthetic valve endocarditis caused by Propionibacterium species: a national registry-based study of 51 Swedish cases. Eur J Clin Microbiol Infect Dis 37:765–771. doi: 10.1007/s10096-017-3172-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3. Download jcm.00341-23-s0001.docx, DOCX file, 0.03 MB (29.1KB, docx)