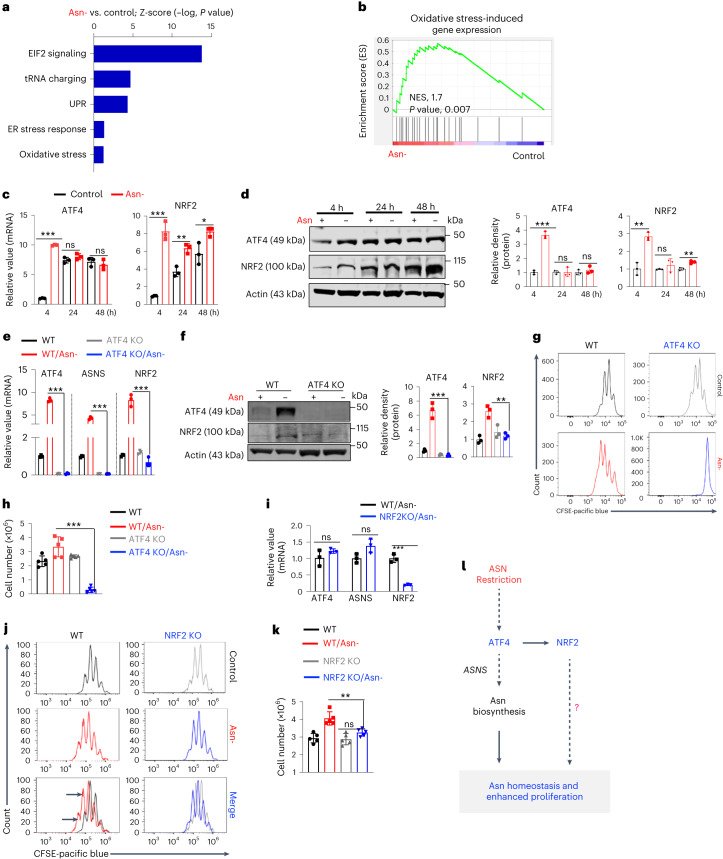
Fig. 4. Asn restriction promotes CD8+ T cell proliferation by enhancing ATF4 and NRF2 expression.
a, Differently expressed gene signatures were determined by the IPA of RNA-seq (cells collected 4 h after activation) and listed according to their z-score (n = 3 experimental replicates). Data are representative of one experiment. ER, endoplasmic reticulum. b, Oxidative stress-induced gene sets were analyzed by gene set enrichment analysis (GSEA). NES, normalized enrichment score. c,d, CD8+ T cells were activated in a medium with or without Asn for the indicated time. mRNA levels and protein levels of indicated molecules were determined by qPCR (c) (n = 3 experimental replicates; ATF4: P < 0.0001, P = 0.2149 and P = 0.4741 for 4 h, 24 h and 48 h, respectively; NRF2: P = 0.0003, P = 0.0039 and P = 0.0303 for 4 h, 24 h and 48 h, respectively). Immunoblot and its quantification were done by ImageJ (v1.53T) (d) (ATF4: P < 0.0001, P = 0.9668 and P = 0.4 for 4 h, 24 h and 48 h, respectively; NRF2: P = 0.007, P = 0.3771 and P = 0.0034 at 4 h, 24 h and 48 h, respectively). Data are representative of three independent experiments. e,f, CD8+ T cells with the indicated genotype and Asn status were collected 4 h after activation. The level of indicated mRNA and protein was determined by qPCR (e) (n = 3 experimental replicates; P < 0.0001, P < 0.0001 and P = 0.0002 for ATF4, ASNS and NRF2, respectively) and immunoblot (f). Immunoblot was quantified by ImageJ (v1.53T) (P = 0.0006 for ATF4 and P = 0.0029 for NRF2). Data are representative of three independent immunoblots. g,h, CD8+ T cells from the indicated genotype were activated in a medium with or without Asn for 72 h. Cell proliferation (g) was determined by CFSE staining, and the cell number was determined by a cell counter (h) (n = 5 experimental replicates; P < 0.0001). Data are representative of four independent experiments. i, The level of the indicated mRNA in cells collected 4 h after activation was determined by qPCR (n = 3 experimental replicates; P = 0.2247, P = 0.0583 and P = 0.0004 for ATF4, ASNS and NRF2, respectively). Data are representative of two independent experiments. j,k, CD8+ T cells from the indicated genotypes were activated in a medium with or without Asn for 72 h. Cell proliferation was determined by CFSE staining (j), and cell number was determined by a cell counter (k) (n = 5 experimental replicates; P = 0.0032). Data are representative of four independent experiments. Error bars represent mean ± s.d. *P < 0.05; **P < 0.01; ***P < 0.001. Statistical differences were determined by paired two-tailed Student’s t-test with a threshold of P < 0.05 (a) and unpaired two-tailed Student’s t-test (c–f, h, i and k). l, The conceptual model of ATF4-dependent NRF2 expression enables robust proliferation of CD8+ Teff cells under Asn restriction.