Summary
Environmental DNA (eDNA) research holds great promise for improving biodiversity science and conservation efforts by enabling worldwide species censuses in near real-time. Current eDNA methods face challenges in detecting low-abundance ecologically important species. In this study, we used isothermal recombinase polymerase amplification (RPA)-CRISPR/Cas detection to test Ctenopharyngodon idella. RPA-CRISPR-Cas12a detected 6.0 eDNA copies/μL within 35 min. Ecologically rare species were identified in the Three Gorges Reservoir Area (TGRA) using functional distinctiveness and geographical restrictiveness, with seven fish species (9%) classified as potentially ecologically rare including three species in this investigation. RPA-CRISPR/Cas12a-FQ outperformed high-throughput sequencing (HTS) and qPCR in detecting low-abundance eDNA (AUC = 0.883∗∗). A significant linear correlation (R2 = 0.682∗∗) between RPA-CRISPR/Cas12a-FQ and HTS quantification suggests its potential for predicting species abundance and enhancing eDNA-based fish biodiversity monitoring. This study highlights the value of RPA-CRISPR/Cas12a-FQ as a tool for advancing eDNA research and conservation efforts.
Subject areas: Ecology, Biological sciences, Molecular biology
Graphical abstract

Highlights
-
•
RPA-CRISPR/Cas12a system is established to sensitively monitor fish from eDNA
-
•
RPA-CRISPR/Cas12a-FQ technique can detect 6.0 copies of fish eDNA with a short time
-
•
RPA-CRISPR/Cas12a-FQ has more promise for rare species identification from eDNA
-
•
CRISPR/Cas12a system is firstly applied for ecological fish monitoring
Ecology; Biological sciences; Molecular biology
Introduction
Biodiversity is rapidly declining at an accelerating rate for most biological groups.1,2 As of 2021, 26,500 species (27% of all assessed species) are listed as endangered on the International Union for Conservation of Nature (IUCN) Red List. Further, aquatic ecosystems are among the most severely altered ecosystems due to pressures from land use, climate change, aquatic invasive species, and water pollution.3,4 Therefore, the development of methods for monitoring biodiversity and tracking and protecting endangered species promptly is urgent. Traditional species monitoring methods rely on sightings and trapping, such as dredging/trawling, gill netting, poisoning, suction sampling, and seine netting.5 These methods can lead to a substantial waste of manpower and material resources and may potentially harm the species of interest, especially endangered species.6,7 Moreover, little can be done about organisms present in low abundance.8
Environmental DNA (eDNA) has emerged as a powerful tool for characterizing biodiversity and detecting changes in ecosystems, due to recent advancements in nucleic acid extraction and detection technology. By capturing and analyzing DNA present in a range of terrestrial and aquatic environments, such as cave sediments, surface soils, and water from lakes, streams, and oceans, eDNA provides a non-invasive method for identifying and tracking the presence of species in the environment.9,10,11,12 Currently, the most commonly used eDNA method involves extracting DNA from various environmental samples (such as air, water, and soil) and analyzing it using a targeted or passive approach. The targeted approach uses species-specific primers with quantitative PCR (qPCR), conventional PCR (PCR), and digital droplet (dd) PCR to detect the presence-absence and estimate the abundance of single species.13,14,15,16 Subsequently, these analytical methods have increasingly been applied to the survey of macro-organisms, particularly in aquatic species.17 However, they are required specific PCR equipment and skilled technicians.
With the advent of next-generation sequencing (NGS), eDNA sequencing offers new opportunities for this process by providing data on the variety, geographical information, and potentially the abundance of species, enabling greater ecosystem protection.18,19 The eDNA metabarcoding method has the advantage of high-throughput detection without the need for developing multiple species-specific markers. However, a critical limitation of this method is its dependence on DNA barcode reference sequence libraries20 and complete DNA,21 which can be incomplete or inaccurate, especially for rare or poorly studied species.
The advent of a CRISPR-based nucleic acid rapid detection platform has recently introduced novel concepts and technological underpinnings for surpassing the limitations of conventional detection methods. This platform that combines nucleic acid pre-amplification with CRISPR-Cas enzymology to specifically identify target DNA or RNA sequences is known as specific high-sensitivity enzymatic reporter unlocking (SHERLOCK).22 Another analogous nucleic acid monitoring tool, the DNA endonuclease-targeted CRISPR trans reporter (DETECTR),23 offers a rapid (approximately 30 min), cost-effective, and accurate CRISPR-Cas12-based lateral flow detection approach that has been employed for viral infection detection. Notably, both SHERLOCK and DETECTR diagnostic tools demonstrate sensitivity and specificity comparable to traditional PCR-based methods, while circumventing the need for complex equipment and are estimated to have low costs.
In this study, with reference to DETECTR, we combined the isothermal recombinase polymerase amplification (RPA) amplification technique with CRISPR/Cas12a and fluorescence detection technology to establish a highly sensitive RPA-CRISPR/Cas12a technique for quantitative detection of rare fish species, and applied this method for accurate in situ detection in the Three Gorges Reservoir Area (TGRA), China. The Three Gorges Reservoir (TGR) holds the distinction of being the largest man-made reservoir globally. The fish resources in the TGRA are vital for biodiversity, food security, and economic development. Compared to previous detection methods, we have more accurately assessed the distribution of ecologically rare fish of the TGRA. Due to the irreplaceable contribution of rare fish species to ecosystem function and the difficulty of detecting their low abundance, this method highlights its invaluable significance for assessing the health of regional ecosystems. To the best of our knowledge, there exists no previous research on the monitoring of ecologically rare fish through the RPA-CRISPR technique in the water.
Results
Species identification via CRISPR/Cas12a-mediated eDNA analysis
The RPA-CRISPR technique was utilized to determine the mtDNA within potentially ecologically rare species of TGRA in field samples collected. To determine if this approach can accurately detect target fish eDNA in wild water bodies, we first validated it with grass carp (Ctenopharyngodon idella), a common species in the TGRA, and the overall concept for the assay is described in Figure 1B.
Figure 1.
Sketch map of Three Gorges Reservoir Area (TGRA) showing the sampling sites and overview of the RPA-CRISPR-Cas12a detection
(A) A total of 28 surface water samples were collected in August and December 2021.
(B) Water samples were collected and filtered before DNA extraction. Target DNA is amplified using the RPA before CRISPR-Cas12a mediated fluorescence detection of the target using a fluorophore quencher (FQ)-labeled ssDNA probe. PAM, protospacer adjacent motif.
To select the crRNA targeting region, the whole mitochondrial sequences of C. idellus, Hypophthalmichthys molitrix, and Hypophthalmichthys nobilis were aligned and visually screened for sites in which only C. idellus included the Cas12a-specific PAM site, which was absent in H. molitrix and H. nobilis (Figures 2A and 3A). We first tested the ability of the designed crRNAs to mediate the homeopathic cleavage activity of the CRISPR/Cas system. The 100 ng of either pUC19 target DNA or PCR products of ND2 in C. idellus complexes were incubated in a 20 μL reaction of the CRISPR/Cas12a system. Clear DNA cleavage was observed at 5 min; with the prolongation of the reaction time, the substrates gradually decreased and the products gradually increased. After 30 min, nearly all of the substrate (pUC19 target DNA or PCR products) was cleaved (Figures 2B and 2C).
Figure 2.
ND2 gene fragment of C. idella was digested with CRISPR/Cas12a system
(A) Location of target binding sequence. The dsDNA cleavage activity of CRISPR/Cas12a system in the plasmid (B) and amplification products (C).
Figure 3.
ND2 gene fragments of C. idella, H. molitrix, and H. nobilis were digested with CRISPR/Cas12a-FQ system
(A) Alignment of crRNA targeting site in C. idella, H. molitrix, and H. nobilis showing PAM sequence, base pair differences, and polymorphic bases.
(B) Fluorescence detection of CRISPR-fluorescence assay to distinguish C. idella ND2 gene and its homologous sequences at 15 min.
(C) Reaction kinetics of CRISPR-fluorescence to distinguish C. idella ND2 gene and its homologous sequences. C. idella recombinant plasmid (D) and whole genomic (F) were detected with the RPA-CRISPR/Cas12a-FQ.
(E) The fluorescent signal analysis of synthetic C. idella DNA with serial dilution. Error bars are mean ± standard deviation, where n = 3. The threshold value is 3 × standard deviation of background fluorescence (samples with no DNA template added).
Using recombinant copies of their respective DNA sequences, we examined whether C. idellus Cas12a-crRNA could discriminate between C. idellus and the closely related species H. molitrix and H. nobilis. C. idellus ND2 gene fragment plasmids and homologous segments from closely related species (H. molitrix and H. nobilis) were treated with Cas12a-crRNA targeting the C. idellus fragment and a single-stranded DNA (ssDNA FQ) reporter. The reporter should only emit a signal upon target recognition and subsequent collateral cleavage by Cas12a nuclease. We demonstrate that our C. idellus-specific Cas12acrRNA emits a fluorescence signal exclusively in the presence of recombinant C. idellus mtDNA and not DNA from closely related species (Figures 3B and 3C).
RPA-CRISPR/Cas12a-FQ technique can detect as little as 6 copies/μL recombinant DNA
To test whether this technique could eventually be applied to the determination of low-abundance species, especially rare species, the different copies of recombinant ND2 were prepared as samples, including 0, 6 × 100, 3 × 101, 6 × 101, 3 ×102, 6 ×102, 3 × 103, 6 × 103, 3 × 104, 6 × 104, 3 × 105, and 6 × 105. The lower limit of detection for the RPA-CRISPR/Cas12a-FQ assay was approximately 6 copies per μL (Figures 3D and 3E). It was necessary to further assess the sensitivity and specificity of this assay in a more complicated genomic setting, i.e., utilizing the C. idellus whole genome as a template as opposed to a simple recombinant plasmid. Using DNA isolated from C. idellus tissue, we demonstrate that RPA coupled to CRISPRCas12a detects C. idellus genomic DNA at concentrations as low as 10−5 ng/μL. This suggests that the assay performance in a whole-genome context was sensitive for C. idellus (Figure 3F).
RPA-CRISPR/Cas12a-FQ technique can distinguish the presence or absence of ecologically rare species from eDNA samples by the HTS and qPCR
To ensure the accuracy of identification at the species level, ASVs with 99% similarity to the 12S rRNA database of fish in the TGRA will be retained. Finally, a total of 76 fish were identified, of which 57 species were identified in summer samples and 61 species in winter samples (Table S3). To reduce the impact of batch effects, we solely use the identification findings of summer samples as the data basis for the subsequent selection of ecologically rare species.
We obtained a distribution based on (1) the geographical range (sampling sites) of species compared to the geographic extent (sampling sites) of all species in the TGRA pool and (2) the functional distinctiveness of species trait values relative to the other species of the TGRA pool (the average functional distance of a species to all the others). Seven fish (9%) were defined as potentially ecologically rare (i.e., both functionally and geographically), while four (5%) were ecologically common (Figure S1, and Table S4).
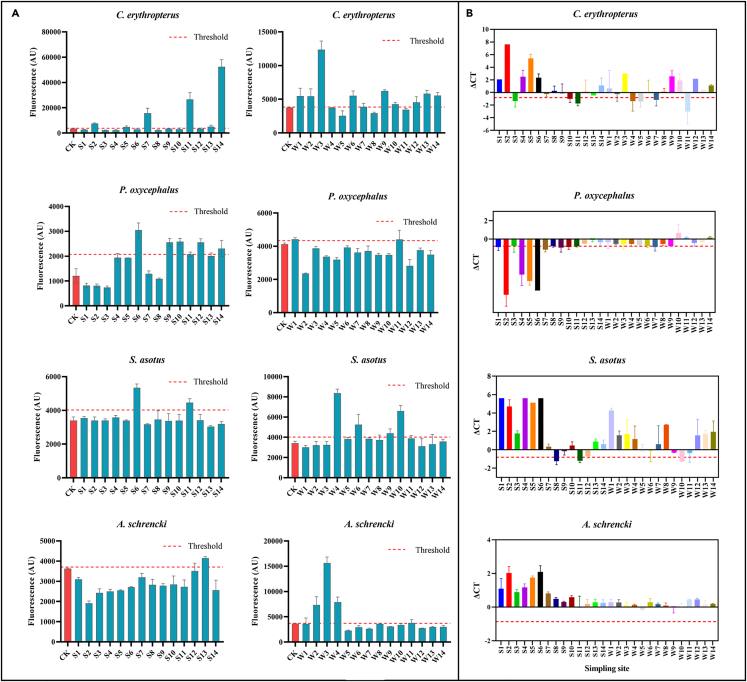
The next test of the assay is to evaluate its applicability to eDNA samples taken in the field, which may contain amplification inhibitors and consist of a substantial proportion of degraded DNA,24 particularly those of ecologically rare species, hence posing additional obstacles. Here, we used eDNA extracted from TGRA samples as templates and previously identified common species C. dellus and ecologically rare species P. oxycephalus, C. erythropterus, and S. asotus using qPCR (Figures 4G and 7B) and HTS of the 12S rRNA gene (Table S3). Within a short-time frame (< 2 h), target species were successfully detected from eDNA extracted from 28 water samples by using the RPA-CRISPR/Cas12a-FQ technique (Figures 4A–4F and 7A). In this study, the existence of a target species in eDNA extracted from water samples was deemed when at least two of three approaches (RPA-CRISPR/Cas12a-FQ, HTS, and qPCR) simultaneously detected the species (Figure 5). As results showed, ROC curves for these 3 methods distinguished positive from negative with all of them had AUC values larger than 0.75, which AUC with the RPA-CRISPR/Cas12a-FQ (0.883) was extremely significantly greater than AUC with HTS (0.804, p value = 0.0066) and qPCR (0.786, p value = 0.0016) (Figure 6A). But there were no substantial differences in the identification effect of HTS and qPCR. Notably, using the RPA-CRISPR/Cas12a-FQ provided up to a positive detection rate of 91.23% (52/57) and a negative detection rate of 85.46% (47/55) (Figure 5). Besides, we monitored the distribution of the unique and rare species of Yangtze River sturgeon during the summer and winter, but this species was not detected in the water sample eDNA of TGRA (Figure 7), indicating that the HTS of the 12S rRNA gene detection method may not be suitable for the detection of all species, particularly rare species. These all indicate that the RPA-CRISPR/Cas12a-FQ has a promising future as an eDNA detection tool. By tracking the distribution of species, particularly ecologically rare species, this detection method has the potential to become one of the most effective instruments for monitoring biodiversity in the natural environment.
Figure 4.
eDNA monitoring of C. idella in the TGRA of Chongqing, China
The dynamics of C. idellus eDNA in summer (A) and winter (B) samples using a microplate reader. Fluorescence measurements were taken every 1 min for 1 h at 37°C. The distribution levels for summer (C) and winter (D) C. idellus in the TGRA using the RPA-CRISPR/Cas12a-FQ detector. The fluorescent signal analysis of C. idella eDNA with summer (E) and winter (F).
(G) The distribution levels for summer and winter C. idellus in the TGRA using qPCR. ΔCt = ΔCt (target) – ΔCt (Control). A Ct value less than 38 is considered valid. S1–14 are the summer samples and W1–14 are the winter samples as mentioned previously. Error bars are mean ± standard deviation, where n = 3. The threshold value is 3 × standard deviation of background fluorescence (samples with no DNA template added).
Figure 7.
The distribution levels of rare fish
(A) The distribution levels for summer and winter rare fish in the TGRA using RPA-CRISPR/Cas12a-FQ detector.
(B) The distribution levels for summer and winter rare fish in the TGRA using qPCR. ΔCt = ΔCt (target) – ΔCt (Control) mean. A Ct value less than 38 is considered valid. S1–14 are the summer samples and W1–14 are the winter samples as mentioned previously. Error bars are mean ± standard deviation, where n = 3. The threshold value is 3 × standard deviation of background fluorescence (samples with no DNA template added).
Figure 5.
Statistics of test results by RPA-CRISPR/Cas12a-FQ, HTS, and qPCR
Green represents positive test results, and black represents negative test results.
Figure 6.
Performance analysis of CRISPR/Cas12a system identification of ecologically rare fish
(A) ROC curves of different methods for eDNA detection.
(B) The standard curve for fish target DNA recombinant plasmids included 0, 5.5 × 100, 5.5 × 101, 5.5 ×102, 5.5 × 103, 5.5 × 104, and 5.5 × 105 copies/μL.
(C) Heatmap of eDNA abundances as measured by the RPA-CRISPR/Cas12a-FQ and HTS.
(D) Correlation (Pearson) between eDNA abundances measured by the RPA-CRISPR/Cas12a-FQ and HTS. (∗∗ indicates p-value < 0.05 and ns indicates not significant).
Species abundance can be estimated by absolute quantification using the RPA-CRISPR/Cas12a-FQ technique
We further tested the quantitative ability of Cas12a. Indeed, this work has already been used to monitor the abundance of Staphylococcus aureus in the Jialing River of Chongqing, China.25 Here, the gene fragments for ND2, ND1, D loop, and ND2 were derived from the P. oxycephalus, C. erythropterus, S. asotus, and C. idella, respectively, were synthesized and constructed into a pUC57 vector. To collect quantitative information on the CRISPR/Cas12a detector system, various copies of recombinant plasmid were prepared as samples, including 0, 5.5 × 100, 5.5 × 101, 5.5 ×102, 5.5 × 103, 5.5 × 104, and 5.5 × 105. Next, we separately constructed standard curves for the four species (Figure 6B). To avoid false positives caused by the different sensitivities of HTS and CRISPR/Cas12a-FQ approaches to eDNA in water samples from impacting the analysis results, only samples detected positive by both HTS and CRISPR/Cas12a-FQ were used in the subsequent analysis (Figure 6C). The abundance of eDNA from each fish species as measured by the RPA-CRISPR/Cas12a-FQ was highly correlated with its relative abundance as measured by HTS (Figure 6D). The correlations were significant (p value = 0.003) with R2 values of 0.682. The clear significant linear correlation between quantified abundances and relative abundances produced by the RPA-CRISPR/Cas12a-FQ and HTS, respectively, indicates that RPA-CRISPR/Cas12a-FQ can be used to measure the concentration of eDNA in samples and estimate species abundance.
Discussion
eDNA methods have been considered as an alternative to traditional monitoring for investigating the distribution of aquatic organisms.26 Numerous studies have reported the use of eDNA to monitor biodiversity in freshwater and marine ecosystems.27,28 However, developing easy, specific, and sensitive on-site assays are still required, especially for lower-abundance rare species, to expand the applications of eDNA. Recently, a field-portable, sensitive fluorimeter chamber was built and successfully applied to Escherichia coli detection. Real-time monitoring of species that are both geographically restricted and functionally different, i.e., possessing rare features and functions, is of the utmost importance due to their extinction risk and possible contribution to the local ecosystem’s functioning.29,30 To achieve this purpose, we first developed the RPA-CRISPR/Cas12a-FQ identification technology and tested the technical performance of the TGRA common species C. idella, which can identify the presence or absence of a field sample with relative ease. We conducted a comprehensive investigation of ecologically rare fish species in part of the TGRA using this method. Furthermore, our novel detection method has demonstrated higher accuracy than previous methods in detecting low-copy rare fish species through comparison. The RPA-CRISPR/Cas12a can be used for accurate field sample detection with a sensitivity of 6.0 copies eDNA per reaction (Figures 3D and 3E), and the entire detection process takes only 35 min. This provides a powerful tool for real-time detection of ecological health and species diversity.
As reported, before detecting a small number of target fragments from eDNA, amplification technologies must be employed.23 The isothermal amplification method RPA may accomplish exponential DNA molecule amplification at a single temperature, unlike traditional PCR techniques, such as qPCR,31,32 which require high-temperature cycles. We designed and synthesized C. idella-specific primers for RPA and paired them with C. idella Cas12a-crRNA for detection. On C. idella-specific recombinant DNA plasmids and genomic DNA, the additional preamplification step demonstrated sensitivity down to 6.0 copies/μL and 10−5 ng/μL level, respectively. Almost consistent with the previous results obtained by employing the technique on Salmo salar.33
To investigate further whether the RPA-CRISPR/Cas12a-FQ can efficiently detect the eDNA of rare species in samples, we computed the ecological rarity of TGRA fish based on the HTS data to select potential functionally rare species for experimental verification. Compared to the ROC curves of the HTS, RPA-CRISPR/Cas12a-FQ, and qPCR detection results of eDNA samples demonstrated that the RPA-CRISPR/Cas12a-FQ has more promise for rare species identification from eDNA.
Previous research has revealed a correlation between biomass and eDNA density in natural or laboratory environments.34 Therefore, this study also explored and experimented with the quantitative aspects of CRISPR technology, and found that the quantitative results of CRISPR technology can reflect the stock of eDNA in environmental samples to a certain extent. However, the quantitative aspects of this assay need to be further explored to obtain more accurate quantitative results, which may involve fidelity processing of eDNA samples35 and optimization of the concentrations of all molecular reagents at each step of the process, etc. Despite the need for additional research on the quantitative components of this assay, evidence suggests that eDNA is superior to other methods for detecting invasive species since it can detect uncommon organisms, such as invasive species.36 However, the development of the RPA/CRISPR-Cas12a-FQ detection has improved the capabilities and uses of eDNA as a warning system for detection and managing ecologically rare or valuable species and has the potential to be adapted to a biosensor device, thus expanding the use of CRISPR/Cas technology to environmental monitoring.
The monitoring of rare species is an important basis for the protection of species diversity. Since the application of eDNA technology, more and more studies conducted for the discovery, monitoring, risk assessment, and management of rare37 and endangered.38 Currently, the detection of rare fish species using eDNA-based methods is primarily accomplished through qPCR and HTS. The former has high sensitivity but is time-consuming and requires complex instrument equipment, while the latter is limited by the size and accuracy of the reference database and errors in the bioinformatics identification of rare species, leading to significant exaggeration in estimates of biodiversity.39 The RPA-CRISPR/Cas12a detection technology for rare fish established in this study has high detection capability and sensitivity along with reduced time costs in comparison to those of the aforementioned two methods. It is worth noting that high sensitivity often comes with the risk of sample cross-contamination, which poses a greater challenge for the development of on-site monitoring devices for this assay in the future.40
Over the past few decades, species rarity has become a cornerstone of ecological research and conservation strategies.41 Specifically, ecologically rare species deserve more attention due to their low abundance and narrow distribution, making them highly susceptible to extinction.42 While it has long been believed that these rare species make minimal contributions to ecosystem functioning and functional diversity, recent research has overturned this prevailing notion. Ecological systems with species possessing unique combinations of traits can indeed make disproportionate contributions to species diversity within a community or region.43 However, until now, these species have rarely been collectively addressed within the context of the intensifying biodiversity crisis. It is now crucial to engage in multi-site and continuous monitoring of geographically restricted and functionally distinct rare species to address present and future threats. The RPA-CRISPR/Cas12a-FQ technology is well-suited for this task when compared to non-targeted eDNA metabarcoding and the stringent experimental conditions of qPCR. Not only does the technology introduce the concept of regional ecological rare species44 for the first time, but it has also been successfully applied to detect these species in multiple sites throughout the TGRA. Furthermore, this marks the first application of the RPA-CRISPR/Cas12a-FQ technology in monitoring aquatic ecological diversity, providing a critical foundation for future global-scale monitoring of rare species. Moreover, the technology is non-selective toward detected samples and requires only the design of species-specific primers and crRNA, making it valuable for conducting continuous, species-specific organism detection across multiple locations. This technique can be especially useful for monitoring flagship species,45 such as manatees, dolphins, sea turtles, and ospreys in marine environments, as well as invasive species in various aquatic systems. Taken together, the successful application of CRISPR technology in the field of ecological conservation provides both technological and conceptual support for advancing biodiversity monitoring and environmental management.
Limitations of the study
Although this study has shown that RPA-CRISPR/Cas12a-FQ technology can be used to estimate fish species abundance, it is not suitable for absolute quantification of species abundance. This is because it requires further consideration of the quantitative relationship between eDNA and species abundance, as well as the kinetics of Cas12a nuclease. Therefore, additional research is needed to explore the quantitative capabilities of this technology. Furthermore, we observed significant variations in the reverse cleavage efficiency of the CRISPR/Cas system with different crRNA sequences, which can affect the sensitivity of the technology. Therefore, it is necessary to design and screen more crRNA sequences before conducting experiments. Additionally, it is important to note that aerosols generated during the RPA process can cause cross-contamination. Despite implementing measures such as cleaning the experimental bench and equipment with disinfectants and alcohol after each experiment, as well as reducing the reuse of primer pairs within a short period, the presence of false positives cannot be completely ruled out.
Inclusion and divsersity
We support inclusive, diverse, and equitable conduct of research.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| DH5α | Sangon Biotech | Cat#B528411 |
| Biological samples | ||
| Tail fins of C. idellus, H. molitrix and H. nobilis | A local seafood market | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Lba Cas12a (cpf1) | New England Biolabs | Cat#M0653T |
| NEBuffer 2.1 | New England Biolabs | Cat#B7202S |
| DNase/RNase-Free Water | Solarbio | Cat#R1600 |
| Critical commercial assays | ||
| TwistAmp Basic Kit | TwistDx | Cat#TABAS03KIT |
| SYBR® Premix Ex Taq ™ | TaKaRa | Cat#RR420A |
| E.Z.N.A.® Soil DNA Kit | OMEGA | Cat#D5625 |
| AxyPrep DNA Gel Extraction Kit | Axygen | Cat#AP-GX-250G |
| QuantiFluor® dsDNA Sample Kit | Promega | Cat# PROE2671 |
| Illumina HiSeq | BIOZERON Biotechnology Co., Ltd | N/A |
| Deposited data | ||
| Raw and analyzed data | This study | BioProject ID: PRJNA881760 |
| Oligonucleotides | ||
| Sequence of crRNA, see Table S1 | This study | N/A |
| Primers for qPCR, see Table S2 | This study | N/A |
| Primers for RPA, see Table S2 | This study | N/A |
| HEX-TTATT-BHQ1 | This study | N/A |
| Recombinant DNA | ||
| mtDNA fragments from C. idella, P. oxycephalus, S. asotus, C. erythropterus and A. schrencki was cloned into a pUC19 vector. | This study | N/A |
| Software and algorithms | ||
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com/ |
| pROC v1.18.0 | Robin et al.46 | http://expasy.org/tools/pROC/ |
| QIIME2 | Bolyen et al.47 | https://qiime2.org/ |
| funrar v1.3 | Grenie et al.44 | https://rekyt.github.io/funrar/ |
Resource availability
Lead contact
Inquiries and requests for resources should be directed to and will be fulfilled by the lead contact, Prof. De-Sheng Pei (peids@cqmu.edu.cn).
Materials availability
The recombinant plasmid in this study will be made available on request to the lead contact; however, requestor will cover shipping costs. This study did not generate new unique reagents.
Experimental model and subject details
The DH5α strain (Cat#B528411) was purchased from Sangon Biotech (Shanghai).
The strain was sub-cultured using LB agar plates (prepared by dissolving 10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl in 1 L of distilled water, followed by autoclaving at 121°C for 15 minutes). We stored the strain in an aqueous stock solution containing glycerol (20%) and froze it at -80°C.
Method details
Sample collection
The samples used in this study were obtained from August 20 to 23, 2021 (the low-water level with 146 m impoundment) and December 26 to 29, 2021 (the high-water level with 175 m impoundment). During both the low-water level period (146 m water level) and the high-water level period (174 m water level), we collected 28 water samples at the same 14 locations along the Yangtze River (106° 23′-111° 04′ E, 29° 19′-31° 04′N) in the Three Gorges reservoir area (TGRA) (Figure 1A). We employed the same sampling technique in both investigations to guarantee the uniformity of the samples. Briefly, 3 L of each water sample was created by merging 1 L water samples from 3 sites, spaced 10 m upstream and downstream of the site. We filtered 3 L of water sample into 3 aliquots for each water sample, labeled each aliquot, and stored it at -80°C until DNA extraction.
Environmental total DNA and tissue DNA extraction
Following the instruction manual, environmental total DNA and tissue DNA from membrane sediment samples and commercially available Ctenopharyngodon idellus, respectively, were extracted using the E.Z.N.A.® Soil DNA Kit (OMEGA, USA). The extracted DNA was then detected by 1% agarose gel electrophoresis.
12S-rDNA barcode reference database for Chinese freshwater fish
To compile the taxonomy information for fish species in the Yangtze River, we gathered data from various sources such as fishbase.org, NCBI, and Mitofish. The NCBI Taxonomy Browser module was employed to extract the taxid for each fish species. Additionally, the 12S-rDNA sequence for each fish was acquired either by querying the term "[taxid] 12S" or by downloading the sequence from MitoFish
PCR amplification and sequencing
PCR amplification and sequencing of the 12S rDNA using Illumina platforms, which generated 250 bp paired-end reads, were carried out by BIOZERON Biotechnology Co., Ltd. (Shanghai, China). Following the manufacturer’s instructions, total DNA was extracted from membrane sediment samples using the E.Z.N.A.® Soil DNA Kit (OMEGA, USA). The integrity of the extracted total DNA was confirmed by 1% agarose gel electrophoresis. The Tele02 primer pairs (forward primer 5′-AAACTCGTGCCAGCCACC-3′; reverse primer 5′-GGGTATCTAATCCCAGTTTG-3′) were designed and evaluated in silico. These primers targeted fragments of the mitochondrial 12S rRNA gene, ranging from 129 to 209 bp (mean = 167 bp), which were subsequently amplified and used for metabarcoding purposes in this study.48 Each sample was subjected to the following PCR conditions: initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 45 s, and a final extension step at 72°C for 10 min. Each sample was replicated three times. The PCR products from the same sample were pooled, and their integrity was verified using 2% agarose gel electrophoresis. Subsequently, the PCR products were purified using the AxyPrep DNA Gel extraction kit (Axygen, USA). The concentration of the resulting library was quantified using the QuantiFluor™-ST blue fluorescence quantification system (Promega, USA). Before sequencing, each library was diluted to a final concentration of 100 pM and combined in equal volumes. The mixed library was sequenced using the NovaSeq 6000 Sequencer (Illumina).
Bioinformatics analyses
The raw data underwent partitioning based on sample barcodes using the barcode splitter, and sequences that did not match any barcode were discarded. A series of quality control processes were conducted using the QIIME tool. Initially, read quality was assessed using fastx_toolkit, and low-quality reads were eliminated using the "split_libraries.py" script with the parameters "-s 20 -w 10 -l 50". Subsequently, chimeras were identified using a de novo approach against the GOLD reference database, and redundant sequences were removed using USEARCH. Following these steps, 53,302,879 clean reads remained. Taxonomic annotation of the sequences was performed using a Naive Bayes classifier trained with a Chinese freshwater fish 12S-rDNA barcode reference library. To achieve a higher resolution of taxonomic annotations and better comparability between investigations, Amplicon Sequence Variants (ASVs) were generated instead of Operational Taxonomy Units (OTUs) with the help of the QIIME2 workflow.47 To minimize the impact of ASVs with extremely low abundance, two filtering criteria were applied: 1) ASVs with fewer than 10 sequences in total across all samples were removed, and 2) ASVs found in fewer than three samples were also removed.
Functional traits
The website fishbase.org (https://www.fishbase.de/) was consulted for data on maximum length, living style, trophic level, and feeding types. According to the FishBase description, "living style" was categorized as "benthopelagic," "demersal," and "pelagic-neritic," while "feeding type" was categorized as "mostly plants/detritus," "primarily animals," and "plants/detritus/animals." If no feeding style information was provided, the nearest relative’s data was used.
Ecological rarity
Using the “funrar” version 1.3 package,44 we then calculated the functional distinctiveness Di of species I in the global functional space, which represents how unique the qualities of a specific species are in comparison to all other species within the same taxon:
where is the total number of species, and is the functional pairwise distance between species and . The scale of the functional distances ranges from 0 to 1. is the mean functional separation between the target species and the remainder of the pool’s species. It illustrates how significantly, on average, the focal species' features diverge from those of the rest of the species pool. When all of the species in the collection have the same trait values (the functional distance between all species is 0), di is equal to 0, and it is equal to 1 when a species' difference from other species is at its greatest.
Using the species matrix of the sampling site, we then computed geographical restrictedness to assess how restricted the distribution of a species is:
Species were divided into three categories based on their values and the quantile divisions of the bivariate space of functional uniqueness against geographic restriction. We classified ecologically rare and ecologically common species as those with values of functional uniqueness and geographical restrictiveness that were greater than 75 percent or less than 25% of the overall species pool of interest, respectively. The functional distinctiveness and geographical restrictiveness values of ecologically average species are, respectively, less than 75% and greater than 25%. With this method, we obtained the ecologically rare species of approach 5% threshold, which is regularly used for the study of the rarity.49,50
Target site identification and primer design
To select target sites for the assay, mitochondrial sequences of Ctenopharyngodon idella, Phoxinus oxycephalus, Chanodichthys erythropterus Silurus asotus, Acipenser schrencki and their congeners or closely related species were obtained from GenBank Release 248 (NCBI). Using the ClustalW alignment tool, the whole mtDNA genomes of the target and closely related species were aligned using multiple sequences. The alignment was visually examined to identify locations where the Cas12a 5′TTTV3′ protospacer adjacent motif (PAM) site51 is unique to the target species. Neighboring PAM sites were searched to guarantee the highest number of mismatches between the target species sequence and closely related species sequences. The final target sites selected reside within the CYTB (A. schrencki), ND2 (P. oxycephalus), ND1 (C. erythropterus), D-loop (S. asotus), and ND2 (C. idella), respectively (Table S1). The identified sequences were used to design the primers and crRNAs. The RPA or qPCR primers were created using the NCBI primer-design tool (https://www.ncbi.nlm.nih.gov/) and their specificity was validated using primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) (Table S2). All oligos including crRNA sequences were purchased from Shanghai Sangon Biotechnology Co., LTD.
Recombinant DNA cloning of target sequences
According to the ND2 gene sequence of mtDNA in C. idella, specific primers were designed via the NCBI primer-design tool, and the primer name was listed in Table S2. The specific 577 bp PCR product within ND2 of C. idella was cloned into a pUC19 vector. Recombinant clones were confirmed by Sanger cycle sequencing using the universal primer M13.
In vitro cleavage assay
The in vitro DNA cleavage assays were performed with either plasmid targets or specific PCR products. The 100 ng either pUC19 target DNA or PCR products complexes were incubated with 1 μL EnLbCas12a (1 μM) (New England Labs), 1 μL crRNA (1 μM), 1 μL NEBuffer2.1(10×), and 6 μL RNase-free water in 20 μL reaction for 20 min at the 37°C. Reactions were inactivated at 85°C for 5 min. The reaction products were run on 1% agarose gels stained with ethidium bromide for production detection.
Isothermal amplification
Target fragments were amplified isothermally with TwistAmp™ Basic Kit using specific primers designed for the test according to the manufacturer’s recommendations. At 39°C, a 25 μL reaction mixture containing 1 μL of DNA sample, 1.2 μL of primer solution (forward primer, reverse primer, 10 mM), 1.2 μL of magnesium acetate (280 mM), 6.6 μL of ddH2O, and 15 μL primer free rehydration buffer was incubated for 20 minutes. Importantly, magnesium acetate was added to the cap and subsequently centrifuged directly into the reaction tube to facilitate simultaneous reaction across all reaction mixtures. The RPA was then introduced for evaluation of CRISPR/Cas12a cleavage.
Cas12a detection reactions
The RPA product (2 μL) was added to 18 μL of the CRISPR-Cas12a reaction mixture containing 1 μL crRNA (1 μM), 1 μL LbCas12a (1 μM) (NEB, Ipswich, UK), 2 μL Reaction NEBuffer 2.1 (10×), 2 μL ssDNA FQ probe sensor (2 μM HEX-sequence-BHQ1), and 12 μL ddH2O (DNase/RNase-free). Then, the reactions (20 μL in a 384-well microplate) were incubated in a fluorescence plate reader (Tecan Infinite® 200 PRO) for up to 60 min at 37°C with fluorescent signals collected every 60 s (ssDNA FQ substrates = λex: 520 nm; λem: 556 nm). For visual detection, all CRISPR reactions were incubated at 37°C for 15 minutes and the intensity of fluorescence was recorded and photographed using a multi-functional ImageQuant reader (AMERSHAM ImageQuant 800, Tecan Company, Switzerland).
qRT-PCR detection
The qRT-PCR of fish eDNA samples was performed with a single-tube PCRs system (TAKARA Biotech Co., Ltd., Dalian, China). The reactions including 10 μL SYBR Premix Ex Taq II (Tli RNaseH Plus) (2×), 0.8 μL primer F and R (10 μM), 0.4 μL ROX Reference Dye Ⅱ, 2 μL of cDNA, and 6 μL of ddH2O. According to the manufacturer’s instruction, the amplification procedure was an initial denaturation step of 95°C for 30 s, followed by 45 cycles of 95°C for 5 s, 60°C for 30 s, and 60°C for 35 s. The fluorescence signal was collected at the 60°C annealing extension per cycle.
Quantification and statistical analysis
The fluorescence value was calculated by subtracting the background fluorescence obtained in the absence of target DNA. The fluorescence threshold was determined as three standard deviations above the background fluorescence observed in reactions without target DNA (n=3). It is important to note that background noise may be present in the sample due to incomplete quenching of the fluorescence reporter. Only fluorescence values exceeding this threshold were considered positive detections of the target. The correlation between the abundance data obtained from high-throughput sequencing (HTS) and CRISPR/Cas was analyzed using Pearson's correlation coefficient, and statistical significance was assessed using a two-tailed test in GraphPad Prism 8 (GraphPad Software, San Diego, CA). The calculation of area under the curves (AUCs) for the three methods (RPA-CRISPR/Cas12a-FQ, HTS, and qPCR) to differentiate between positive and negative samples was performed using the R package "pROC" (version: 1.18.0).46 Standard curves were generated using GraphPad Prism with a linear regression fit and a 95% confidence interval. Asterisks denote the level of statistical significance: ∗, p ≤ 0.05; ∗∗, p ≤ 0.01; ∗∗∗, p ≤ 0.001.
Acknowledgments
We thank the support from the Chongqing Medical University Talent Project (No. R4014 to D.S.P.), Chongqing Postdoctoral Innovation Mentor Studio (X7928 D.S.P.), China-Sri Lanka Joint Research and Demonstration Center for Water Technology, and China Sri Lanka Joint Center for Education and Research, Chinese Academy of Sciences, China.
Author contributions
D.S.P. and X.Y.W. designed the study, X.Y.W. conducted the experiment. L.L., H.H., H.J.J., and L.K.B. analyzed the data. X.Y.W. and D.S.P. wrote the manuscript and revised the paper. D.S.P. provides supervision, project administration, and funding acquisition. This manuscript was approved by all authors.
Declaration of interests
The authors declare no conflict of interest.
Published: August 3, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107519.
Supplemental information
Data and code availability
The sequencing data is available in GenBank within the BioProject number PRJNA881760 with a live link: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA881760. The lead contact can provide any further details necessary upon request for reanalyzing the data presented in this study. All the data produced in this study have been either included in the published paper and its accompanying supplementary materials, or are accessible through the lead contact upon request.
References
- 1.Pimm S.L., Jenkins C.N., Abell R., Brooks T.M., Gittleman J.L., Joppa L.N., Raven P.H., Roberts C.M., Sexton J.O. The biodiversity of species and their rates of extinction, distribution, and protection. Science. 2014;344 doi: 10.1126/science.1246752. [DOI] [PubMed] [Google Scholar]
- 2.Toussaint A., Brosse S., Bueno C.G., Pärtel M., Tamme R., Carmona C.P. Extinction of threatened vertebrates will lead to idiosyncratic changes in functional diversity across the world. Nat. Commun. 2021;12:5162. doi: 10.1038/s41467-021-25293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vörösmarty C.J., McIntyre P.B., Gessner M.O., Dudgeon D., Prusevich A., Green P., Glidden S., Bunn S.E., Sullivan C.A., Liermann C.R., Davies P.M. Global threats to human water security and river biodiversity. Nature. 2010;467:555–561. doi: 10.1038/nature09440. [DOI] [PubMed] [Google Scholar]
- 4.Bernhardt E.S., Rosi E.J., Gessner M.O. Synthetic chemicals as agents of global change. Front. Ecol. Environ. 2017;15:84–90. doi: 10.1002/fee.1450. [DOI] [Google Scholar]
- 5.Bonar S.A., Hubert W.A., Willis D.W. 2009. Standard Methods for Sampling North American Freshwater Fishes. [Google Scholar]
- 6.Snyder D.E. Invited overview: conclusions from a review of electrofishing and its harmful effects on fish. Rev. Fish Biol. Fish. 2003;13:445–453. doi: 10.1007/s11160-004-1095-9. [DOI] [Google Scholar]
- 7.Schmelzle M.C., Kinziger A.P. Using occupancy modelling to compare environmental DNA to traditional field methods for regional-scale monitoring of an endangered aquatic species. Mol. Ecol. Resour. 2016;16:895–908. doi: 10.1111/1755-0998.12501. [DOI] [PubMed] [Google Scholar]
- 8.MAGNUSON J.J., BENSON B.J., MCLAIN A.S. Insights on Species Richness and Turnover from Long-Term Ecological Research: Fishes in North Temperate Lakes1. Am. Zool. 1994;34:437–451. [Google Scholar]
- 9.Pedersen M.W., Overballe-Petersen S., Ermini L., Sarkissian C.D., Haile J., Hellstrom M., Spens J., Thomsen P.F., Bohmann K., Cappellini E., et al. Ancient and modern environmental DNA. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370 doi: 10.1098/rstb.2013.0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schreiber L., Castellanos-Galindo G.A., Robertson D.R., Torchin M., Chavarria K., Laakmann S., Saltonstall K. Environmental DNA (eDNA) reveals potential for interoceanic fish invasions across the Panama Canal. Ecol. Evol. 2023;13 doi: 10.1002/ece3.9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivera S.F., Rimet F., Vasselon V., Vautier M., Domaizon I., Bouchez A. Fish eDNA metabarcoding from aquatic biofilm samples: Methodological aspects. Mol. Ecol. Resour. 2022;22:1440–1453. doi: 10.1111/1755-0998.13568. [DOI] [PubMed] [Google Scholar]
- 12.Yao M., Zhang S., Lu Q., Chen X., Zhang S.Y., Kong Y., Zhao J. Fishing for fish environmental DNA: Ecological applications, methodological considerations, surveying designs, and ways forward. Mol. Ecol. 2022;31:5132–5164. doi: 10.1111/mec.16659. [DOI] [PubMed] [Google Scholar]
- 13.Carlsson J.E., Egan D., Collins P.C., Farrell E.D., Igoe F., Carlsson J. A qPCR MGB probe based eDNA assay for European freshwater pearl mussel (Margaritifera margaritifera L.) Aquat. Conserv. 2017;27:1341–1344. doi: 10.1002/aqc.2788. [DOI] [Google Scholar]
- 14.Baker C.S., Steel D., Nieukirk S., Klinck H. Environmental DNA (eDNA) From the Wake of the Whales: Droplet Digital PCR for Detection and Species Identification. Front. Mar. Sci. 2018;5:133. doi: 10.3389/fmars.2018.00133. [DOI] [Google Scholar]
- 15.McCarthy A., Rajabi H., McClenaghan B., Fahner N.A., Porter E., Singer G.A.C., Hajibabaei M. Comparative analysis of fish environmental DNA reveals higher sensitivity achieved through targeted sequence-based metabarcoding. Mol. Ecol. Resour. 2023;23:581–591. doi: 10.1111/1755-0998.13732. [DOI] [PubMed] [Google Scholar]
- 16.Pont D., Meulenbroek P., Bammer V., Dejean T., Erős T., Jean P., Lenhardt M., Nagel C., Pekarik L., Schabuss M., et al. Quantitative monitoring of diverse fish communities on a large scale combining eDNA metabarcoding and qPCR. Mol. Ecol. Resour. 2023;23:396–409. doi: 10.1111/1755-0998.13715. [DOI] [PubMed] [Google Scholar]
- 17.Yates M.C., Cristescu M.E., Derry A.M. Integrating physiology and environmental dynamics to operationalize environmental DNA (eDNA) as a means to monitor freshwater macro-organism abundance. Mol. Ecol. 2021;30:6531–6550. doi: 10.1111/mec.16202. [DOI] [PubMed] [Google Scholar]
- 18.Beans C. Core Concept: Environmental DNA helps researchers track pythons and other stealthy creatures. Proc. Natl. Acad. Sci. USA. 2018;115:8843–8845. doi: 10.1073/pnas.1811906115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guisan A., Tingley R., Baumgartner J.B., Naujokaitis-Lewis I., Sutcliffe P.R., Tulloch A.I.T., Regan T.J., Brotons L., McDonald-Madden E., Mantyka-Pringle C., et al. Predicting species distributions for conservation decisions. Ecol. Lett. 2013;16:1424–1435. doi: 10.1111/ele.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hebert P.D.N., Cywinska A., Ball S.L., DeWaard J.R. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lacoursière-Roussel A., Dubois Y., Normandeau E., Bernatchez L. Improving herpetological surveys in eastern North America using the environmental DNA method. Genome. 2016;59:991–1007. doi: 10.1139/gen-2015-0218. [DOI] [PubMed] [Google Scholar]
- 22.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taberlet P., Coissac E., Pompanon F., Brochmann C., Willerslev E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 2012;21:2045–2050. doi: 10.1111/j.1365-294X.2012.05470.x. [DOI] [PubMed] [Google Scholar]
- 25.Liu L., Duan J.J., Wei X.Y., Hu H., Wang Y.B., Jia P.P., Pei D.S. Generation and application of a novel high-throughput detection based on RPA-CRISPR technique to sensitively monitor pathogenic microorganisms in the environment. Sci. Total Environ. 2022;838 doi: 10.1016/j.scitotenv.2022.156048. [DOI] [PubMed] [Google Scholar]
- 26.Thomsen P.F., Sigsgaard E.E. Environmental DNA metabarcoding of wild flowers reveals diverse communities of terrestrial arthropods. Ecol. Evol. 2019;9:1665–1679. doi: 10.1002/ece3.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deutschmann B., Müller A.K., Hollert H., Brinkmann M. Assessing the fate of brown trout (Salmo trutta) environmental DNA in a natural stream using a sensitive and specific dual-labelled probe. Sci. Total Environ. 2019;655:321–327. doi: 10.1016/j.scitotenv.2018.11.247. [DOI] [PubMed] [Google Scholar]
- 28.Thomsen P.F., Willerslev E. Environmental DNA - An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015;183:4–18. doi: 10.1016/j.biocon.2014.11.019. [DOI] [Google Scholar]
- 29.Loiseau N., Mouquet N., Casajus N., Grenié M., Guéguen M., Maitner B., Mouillot D., Ostling A., Renaud J., Tucker C., et al. Global distribution and conservation status of ecologically rare mammal and bird species. Nat. Commun. 2020;11:5071. doi: 10.1038/s41467-020-18779-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trindade-Santos I., Moyes F., Magurran A.E. Global patterns in functional rarity of marine fish. Nat. Commun. 2022;13:877. doi: 10.1038/s41467-022-28488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shelton A.O., Kelly R.P., O'Donnell J.L., Park L., Schwenke P., Greene C., Henderson R.A., Beamer E.M. Environmental DNA provides quantitative estimates of a threatened salmon species. Biol. Conserv. 2019;237:383–391. doi: 10.1016/j.biocon.2019.07.003. [DOI] [Google Scholar]
- 32.Coble A.A., Flinders C.A., Homyack J.A., Penaluna B.E., Cronn R.C., Weitemier K. eDNA as a tool for identifying freshwater species in sustainable forestry: A critical review and potential future applications. Sci. Total Environ. 2019;649:1157–1170. doi: 10.1016/j.scitotenv.2018.08.370. [DOI] [PubMed] [Google Scholar]
- 33.Williams M.A., O'Grady J., Ball B., Carlsson J., de Eyto E., McGinnity P., Jennings E., Regan F., Parle-McDermott A. The application of CRISPR-Cas for single species identification from environmental DNA. Mol. Ecol. Resour. 2019;19:1106–1114. doi: 10.1111/1755-0998.13045. [DOI] [PubMed] [Google Scholar]
- 34.Salter I., Joensen M., Kristiansen R., Steingrund P., Vestergaard P. Environmental DNA concentrations are correlated with regional biomass of Atlantic cod in oceanic waters. Commun. Biol. 2019;2:461. doi: 10.1038/s42003-019-0696-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cantera I., Cilleros K., Valentini A., Cerdan A., Dejean T., Iribar A., Taberlet P., Vigouroux R., Brosse S. Optimizing environmental DNA sampling effort for fish inventories in tropical streams and rivers. Sci. Rep. 2019;9:3085. doi: 10.1038/s41598-019-39399-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi S., Sakata M.K., Minamoto T., Masuda R. Comparing the efficiency of open and enclosed filtration systems in environmental DNA quantification for fish and jellyfish. PLoS One. 2020;15 doi: 10.1371/journal.pone.0231718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balasingham K.D., Walter R.P., Mandrak N.E., Heath D.D. Environmental DNA detection of rare and invasive fish species in two Great Lakes tributaries. Mol. Ecol. 2018;27:112–127. doi: 10.1111/mec.14395. [DOI] [PubMed] [Google Scholar]
- 38.Robinson C.V., Uren Webster T.M., Cable J., James J., Consuegra S. Simultaneous detection of invasive signal crayfish, endangered white-clawed crayfish and the crayfish plague pathogen using environmental DNA. Biol. Conserv. 2018;222:241–252. doi: 10.1016/j.biocon.2018.04.009. [DOI] [Google Scholar]
- 39.Flynn J.M., Brown E.A., Chain F.J.J., MacIsaac H.J., Cristescu M.E. Toward accurate molecular identification of species in complex environmental samples: testing the performance of sequence filtering and clustering methods. Ecol. Evol. 2015;5:2252–2266. doi: 10.1002/ece3.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heery B., Briciu-Burghina C., Zhang D., Duffy G., Brabazon D., O'Connor N., Regan F. ColiSense, today's sample today: A rapid on-site detection of beta-D-Glucuronidase activity in surface water as a surrogate for E. coli. Talanta. 2016;148:75–83. doi: 10.1016/j.talanta.2015.10.035. [DOI] [PubMed] [Google Scholar]
- 41.Enquist B.J., Feng X., Boyle B., Maitner B., Newman E.A., Jørgensen P.M., Roehrdanz P.R., Thiers B.M., Burger J.R., Corlett R.T., et al. The commonness of rarity: Global and future distribution of rarity across land plants. Sci. Adv. 2019;5:eaaz0414. doi: 10.1126/sciadv.aaz0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaston K.J. Ecology - Rarity as double jeopardy. Nature. 1998;394:229–230. doi: 10.1038/28288. [DOI] [Google Scholar]
- 43.Dee L.E., Cowles J., Isbell F., Pau S., Gaines S.D., Reich P.B. When Do Ecosystem Services Depend on Rare Species? Trends Ecol. Evol. 2019;34:746–758. doi: 10.1016/j.tree.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 44.Grenié M., Denelle P., Tucker C.M., Munoz F., Violle C. funrar: An R package to characterize functional rarity. Divers. Distrib. 2017;23:1365–1371. doi: 10.1111/ddi.12629. [DOI] [Google Scholar]
- 45.Sergio F., Newton I., Marchesi L., Pedrini P. Ecologically justified charisma: preservation of top predators delivers biodiversity conservation. J. Appl. Ecol. 2006;43:1049–1055. doi: 10.1111/j.1365-2664.2006.01218.x. [DOI] [Google Scholar]
- 46.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C., Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taberlet P., Bonin A., Zinger L., Coissac R. Environmental DNA: For Biodiversity Research and Monitoring; 2018. Environmental DNA: For Biodiversity Research and Monitoring. [Google Scholar]
- 49.Violle C., Thuiller W., Mouquet N., Munoz F., Kraft N.J.B., Cadotte M.W., Livingstone S.W., Mouillot D. Functional Rarity: The Ecology of Outliers. Trends Ecol. Evol. 2017;32:356–367. doi: 10.1016/j.tree.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prendergast J.R., Quinn R.M., Lawton J.H., Eversham B.C., Gibbons D.W. Rare species, the coincidence of diversity hotspots and conservation strategies. Nature. 1993;365:335–337. [Google Scholar]
- 51.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A., et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data is available in GenBank within the BioProject number PRJNA881760 with a live link: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA881760. The lead contact can provide any further details necessary upon request for reanalyzing the data presented in this study. All the data produced in this study have been either included in the published paper and its accompanying supplementary materials, or are accessible through the lead contact upon request.