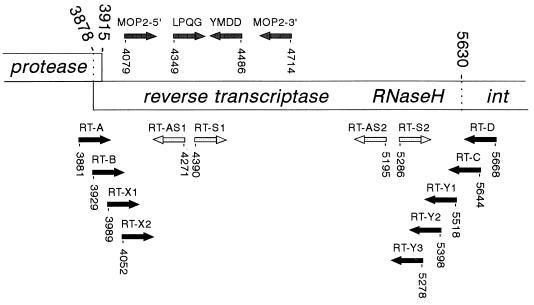
FIG. 1.
Schematic of part of the HERV-K genome encompassing the RT and RNase H domains. This genome structure was originally proposed by Ono et al. for the prototype HERV-K10 element (36), and all nucleotide numbers refer to positions in this genome. The protease gene is in the +1 reading frame compared with RT and ends at stop codon 3915. Thus, a −1 frameshift is expected to occur in the upstream region to fuse the protease and RT reading frames. The integrase enzyme (int) is in frame with the RT gene. The protease cleavage sites, which, by analogy to exogenous retroviruses, will determine the ends of the mature RT enzyme, are not known. The RT primer sets RT-A plus RT-D and RT-B plus RT-C (black arrows) were used in this study to amplify full-length RT genes, and the RT-X1 plus RT-X2 and RT-Y1 through RT-Y3 primers (black arrows) were used to construct N-terminal and C-terminal deletion variants of the RT enzyme (Fig. 7). The LPQG plus YIDD and MOP2-5′ plus MOP2-3′ primer sets shown above the RT gene (grey arrows) have been used previously to amplify internal RT fragments (24, 33). The sense primers S1 and S2 and the antisense primers AS1 and AS2 (open arrows) were used in this study for sequencing of the complete RT gene.