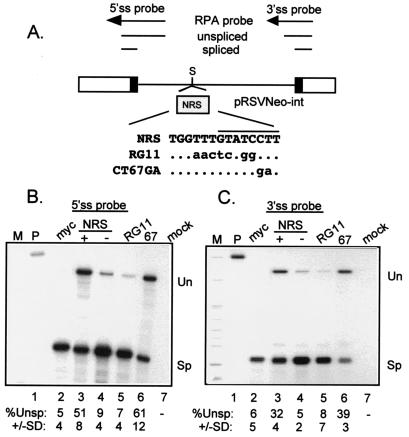
FIG. 3.
The CT67GA NRS mutation does not impair NRS splicing inhibition. (A) Diagram of the pRSVNeo-int construct and RNase protection probes used to measure NRS activity. Open boxes indicate Neo sequences, black boxes represent the small portions of myc exons, and the line denotes the myc intron. Above is a schematic of the RNase protection probes and protected fragments. The shaded box represents NRS fragments that were inserted into the SacII site (S) of the myc intron of pRSVNeo-int. The sequence surrounding the U11 site of the mutant and wild-type NRS fragments is shown. The dots indicate unchanged bases; changes are in lowercase. The U11 site is overlined. (B and C) RNase protection assays on RNA from transfected 293 cells. Total RNA from 293 cells transfected with the indicated constructs was used for RNase protection with 5′ ss (B) and 3′ ss (C) probes (see Materials and Methods), and protected fragments were run on a 4% (B) or 6% (C) denaturing polyacrylamide gels and visualized by autoradiography. M, markers; P, unprocessed probe; myc, pRSVNeo-int with no insert; + and −, sense and antisense orientations of the NRS; RG11 and CT67GA (67) NRS mutants; mock, RNA from mock-transfected cells; Un and Sp, positions of bands representing unspliced and spliced RNA. Below the lanes is the quantitated percent unspliced RNA as determined by PhosphorImager analysis; the data are representative of at least three separate experiments.