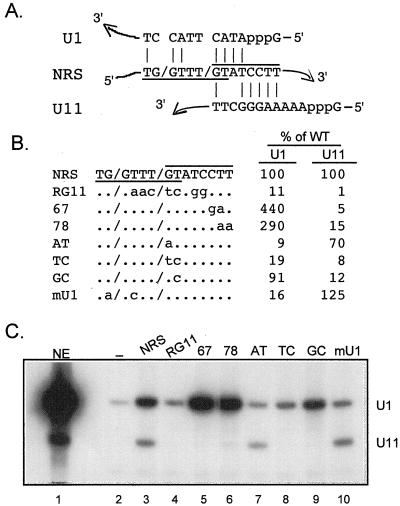
FIG. 4.
A U1 snRNP binding site overlaps the U11 site. (A) Diagram of potential base pairing interactions (vertical lines) of U1 and U11 snRNA with the NRS. The NRS U1 site is underlined; the U11 site is overlined; slashes indicate where splicing would be predicted to occur if the sites were functional. (B) Sequences of NRS and mutants used for affinity selection. To the right, bands in panel C were quantitated with a PhosphorImager, and the intensity of each of the U1 and U11 signals generated by the mutants was compared to that for wild-type (WT) NRS RNA, which was set to 100%. (C) Affinity selection. Equal moles of the indicated biotinylated RNAs were incubated in nuclear extract and affinity selected with streptavidin-agarose beads, and snRNA components of snRNPs associated with the NRS were extracted with phenol-chloroform (see Materials and Methods). The extracted RNA was resolved in a denaturing polyacrylamide gel, electroblotted to a nylon membrane, and hybridized with U1 and U11 antisense riboprobes. Background binding to the beads was determined with nonbiotinylated RNA (−). NE, U1 and U11 snRNA markers extracted from 3 μl of nuclear extract. The positions of U1 and U11 are indicated on the right.