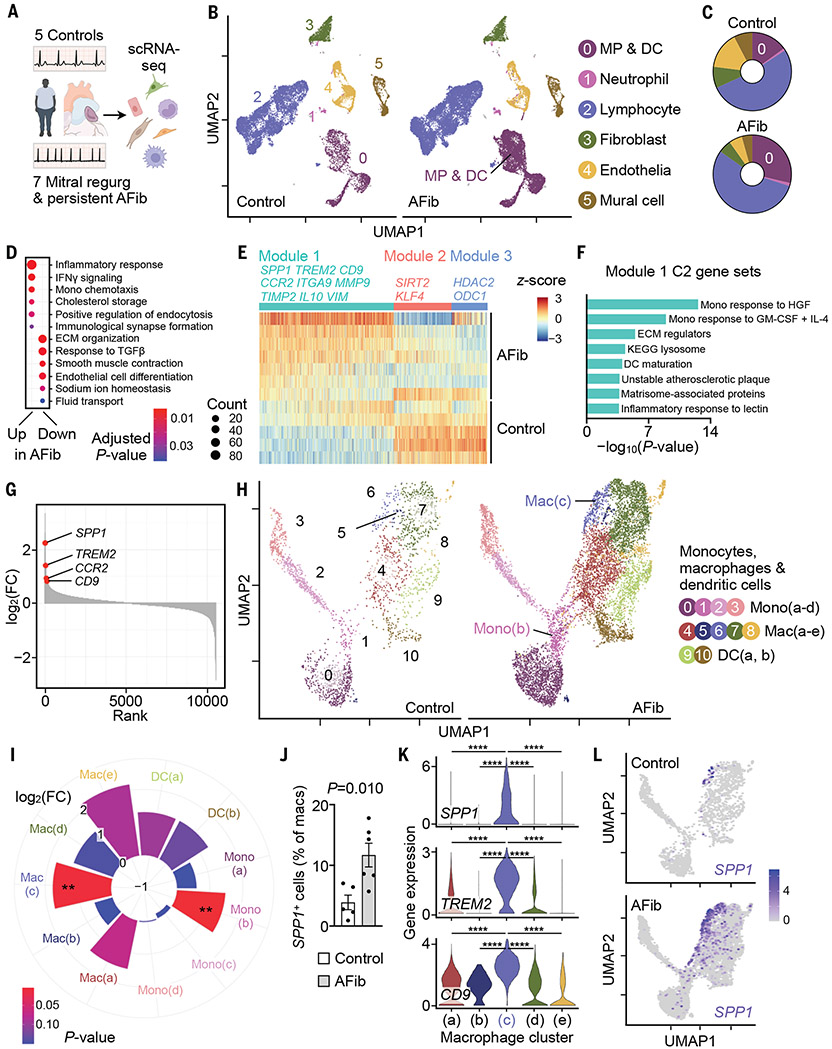
Fig. 1. Single-cell atlas of human atrial disease.
(A) scRNA-seq was performed in five controls and seven patients with AFib. (B) UMAP delineates six major cardiac cell types. (C) Distribution of cell populations. Color code corresponds to (B). (D) Gene ontology biological process (GOBP) gene sets (48, 49) up- or down-regulated in AFib from GSEA of MP/DC cluster. Circle size denotes number of enriched genes. ECM, extracellular matrix. (E) Gene expression in MP/DC cluster represented by z-scores of the log-transformed normalized counts. Rows, samples grouped by disease state; columns, genes in the three gene modules significantly associated with disease state; gene coexpression modules and exemplary genes are labeled. (F) Select MSigDB C2 gene sets (50) overrepresented in module 1 described in (E). GM-CSF, granulocyte-macrophage colony-stimulating factor; HGF, hepatocyte growth factor; IL-4, interleukin-4; KEGG, Kyoto Encyclopedia of Genes and Genomes. (G) Genes in MP/DC cluster ranked according to log2-fold changes [log2(FC)] between seven patients with AFib and five controls. (H) Subclustering of 10,555 MP/DCs from five controls and seven patients with AFib. (I) Difference in abundance of cells belonging to MP/DC subclusters between seven patients with AFib and five control patients. Pie size indicates log2(FC); color (blue, high; red, low) indicates the P value (two-tailed Student’s t test). **P < 0.01. (J) Fraction of SPP1+ macrophages in controls and patients with AFib. Bar graph shows mean ± SEM, n = 5 to 6 per group, two-tailed Student’s t test. (K) Gene expression levels (represented by log-transformed normalized counts) across macrophage clusters; Kruskal–Wallis (P < 2.2 × 10−16) followed by two-tailed Wilcoxon rank sum test. ****Adjusted P = 2 × 10−16. (L) SPP1 expression in MP/DCs from controls and patients with AFib.