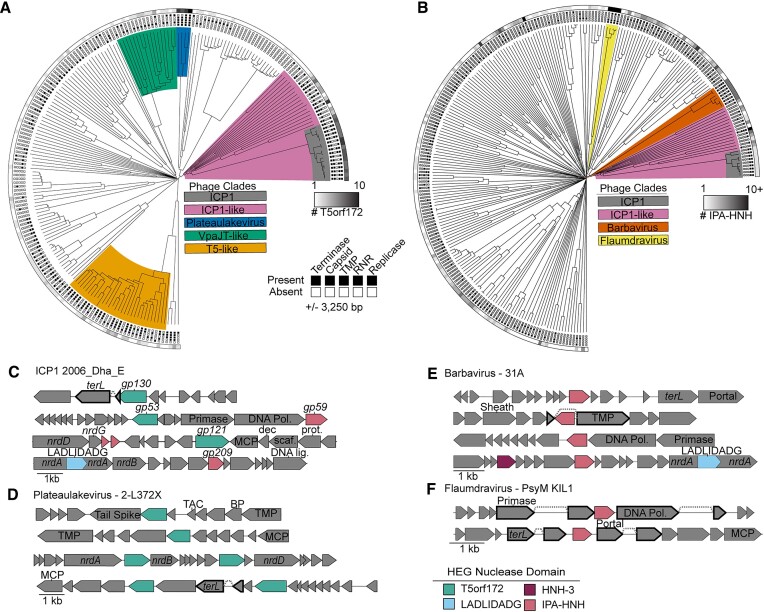
Figure 2.
HEGs cluster near essential genes in diverse phage lineages. (A, B) T5orf172 (A) and IPA-HNH (B) HEGs are often localized in gene neighborhoods that include essential phage genes. Candidate phage genomes containing homologs to ICP1 representative nucleases were identified by PSI-BLAST and the gene neighborhoods containing the ± 3250 bp flanking each predicted HEG present were annotated for predicted function. Phylogenies of phage genomes were generated using the Viptree (34) algorithm to generate a proteomic tree of the phage relatedness based on tBLASTx. Gene neighborhoods including genes predicted to encode a phage terminase, capsid, tape measure protein (TMP), ribonucleotide reductase (RNR), or replicase component were indicated with a black box at the end of the leaf. The number of putative HEGs in each genome is represented by the grayscale bar surrounding the tree, with all phages encoding at least one representative HEG. Clades with a high density of HEGs discussed throughout the manuscript are highlighted accordingly. (C–F) Representative gene neighborhoods for T5orf172 (C, D) and IPA-HNH (C, E, F) HEGs show the co-localization of HEGs with essential phage genes. Validated and predicted exon splicing junctions are indicated by a dashed line connecting two genes, which are outlined in bold. Genes encoded by ICP1 are shown with their gp number, as referenced in Figure 1. Colored boxes next to phage names indicate the phage clade as colored in (A) or (B). Functional predictions for phage genes are based on the Pfam domain predictions using hmmsearch. Domain key: terL = large terminase, DNA Pol = DNA polymerase, nrdA/B/D = ribonucleoside-triphosphate reductase, MCP = major coat subunit, Dec = decoration protein, Scaf = scaffold, Prot = scaffold protease, TAC = tail assembly chaperone, BP = baseplate, TMP = tape measure protein.