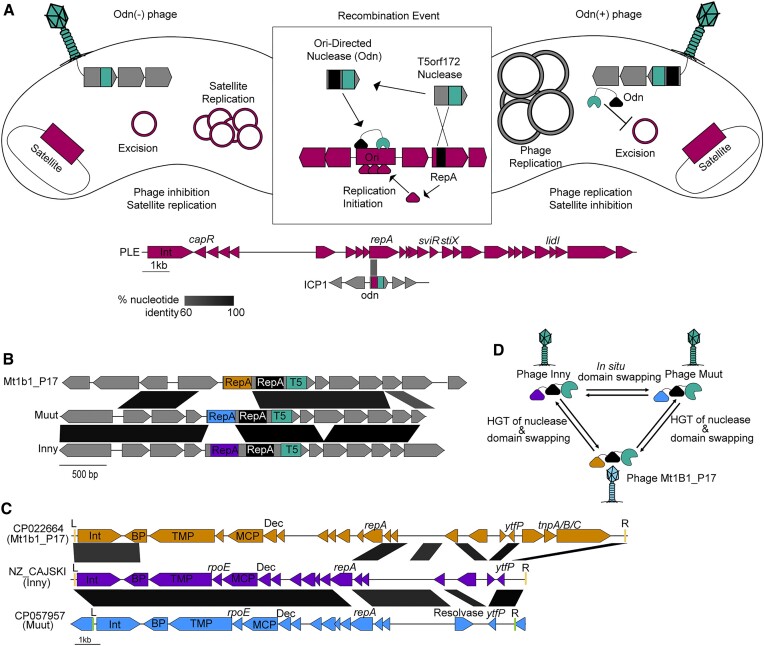
Figure 8.
HEGs are broadly involved in the phage-satellite evolutionary arms race. (A) A representative model of the mechanism of satellite inhibition by Odn. In the absence of Odn, the phage satellite life cycle continues unperturbed, inhibiting phage progeny production. Following recombination to acquire Odn, phages are able to overcome the satellite and viable phages are generated from the infection. The PLE gene graph shows the features of PLE 1 and highlights characterized PLE features with labels above the diagram. (B) Examples of phages that are predicted to encode novel odn genes. RepA_N domains of putative odn genes are colored identically to their corresponding satellite in panel C. Regions of nucleotide identity greater than 60% are shaded accordingly. Each odn homolog encodes a second RepA_N domain without detectable homology to cognate satellite repA genes, which is shown in black. (C) Putative phage satellites that are predicted to be targets of each respective odn homolog indicated in panel B. Gene graphs are colored according to phage genome, which corresponds to the odn RepA domain in panel B. Predicted attL and attR recombinase attachment sites are indicated with red (tRNA integrated) and green (lysR integrated) lines and marked L and R. Genes with a strong functional prediction from HHsearch analysis are labeled with gene names or predicted protein function. Dec = decoration, MCP = major capsid protein, Int = integrase, BP = base-plate hub, TMP = tape measure protein. (D) A schematic of the hypothetical exchanges of domains between related phage odn homologs