Figure 5. Bambu enables the discovery and quantification of highly repetitive genes.
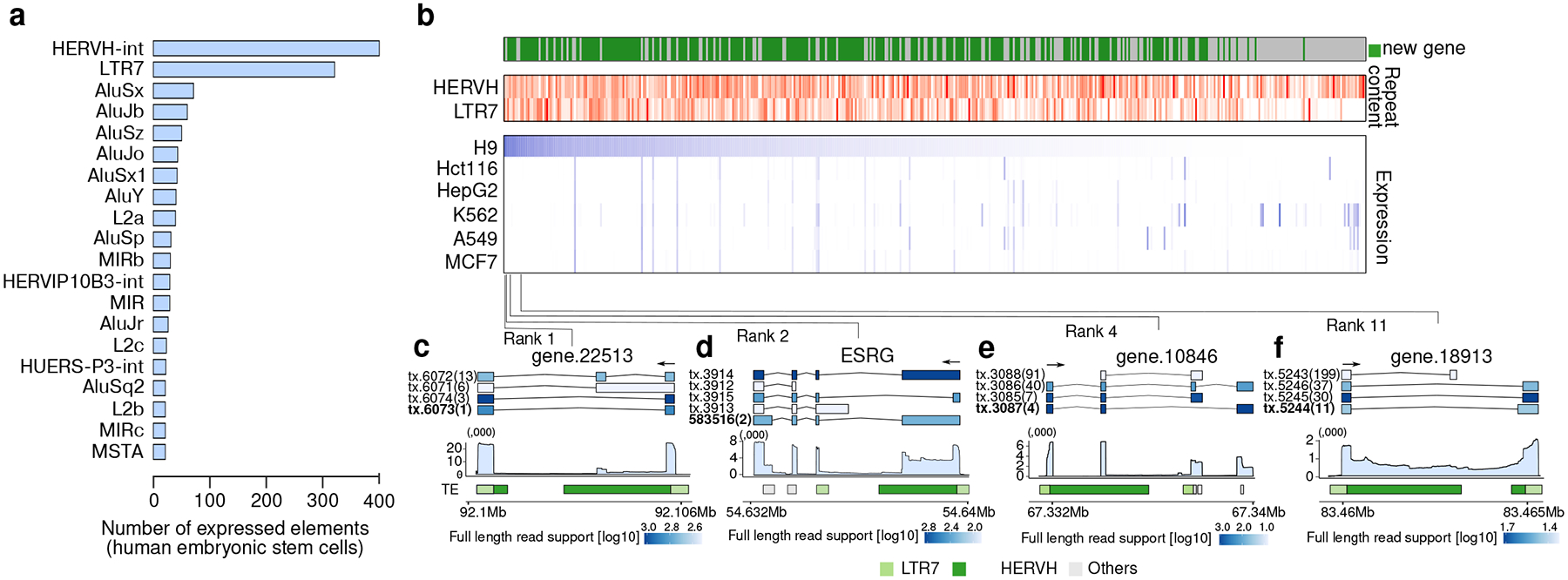
(a) Repeat families ranked by the number of expressed elements identified in the human embryonic stem cell cell line (H9) (b) Overview of the top 100 expressed novel and annotated transcripts that overlap with the HERVH-LTR7 retrotransposon in hESC cell line. For novel transcripts, we only include those with high overlap in novel exons. Top: Fraction of overlap with HERVH and LTR7. Bottom: Expression estimate in H9 hESCs and in the 5 SG-NEx cancer cell lines (c-f) Illustrations of highly expressed transcripts in H9 hESCs that originate from HERVH-LTR7 repeats that show distinct splicing patterns and transcript sequences. Top: transcript annotation colored by estimated full-length read support, with ranks in expression highlighted inside the bracket. Middle: mean read coverage for the specified genomic ranges for each selected gene. Bottom: show repeat masker colored by HERVH (green), LTR7 (light green), and all other repeat types (light grey).
