Figure 1.
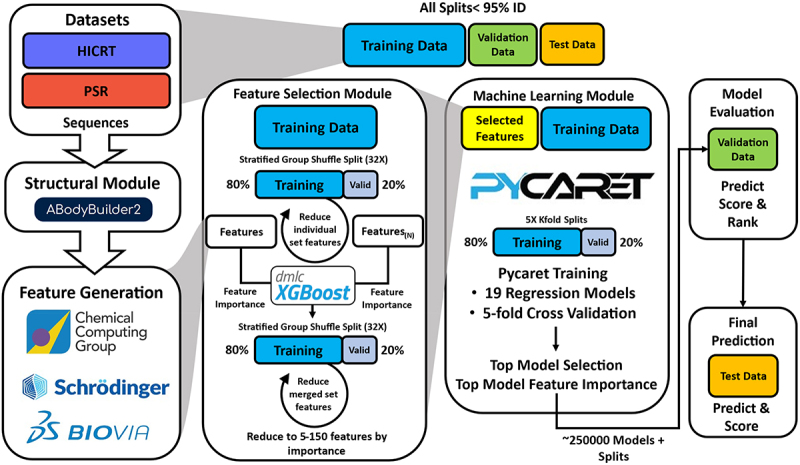
Schematic of machine learning workflow in this study.
For each bioassay endpoint in the Datasets category, the data are split into training validation test sets, ensuring low sequence identity, and representative assay variation in the validation set. Fv sequences are modeled using ABodyBuilder2 and features are generated using three popular software packages. Individual or multiple feature sets are trained with XGBoost regression models using 32 grouped splits and reduced individually to X features (X is a hyparameter 5-150). Multiple reduced feature sets are then combined, resubmitted, and reduced again to X features. The top features selected by XGBoost are submitted with the training set to a PyCaret workflow and trained on 19 regression models. For each X value, the top five models are then tested for prediction and scored on the validation set for ten random seeds. 5000 cycles (250K) models are evaluated and ranked on the validation data. The top performing model on the validation data is confirmed by prediction on the test set.
