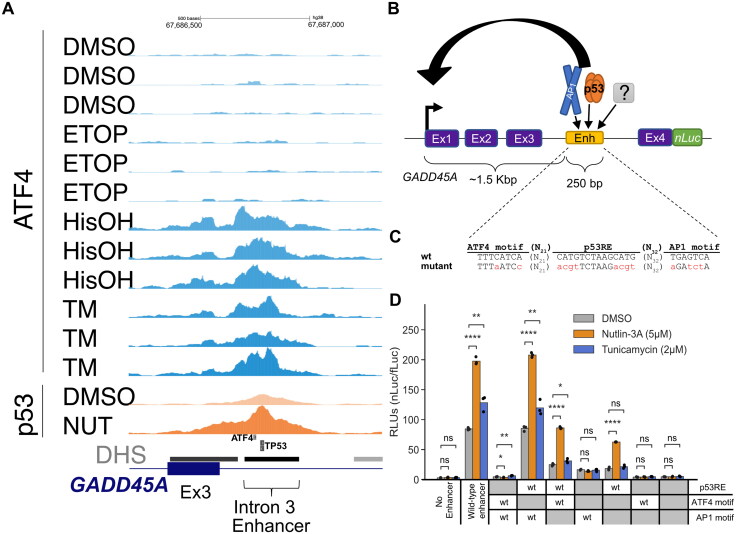
FIG 7.
GADD45A as a reporter system to study DDR- and ISR-dependent enhancers. (A) Genome browser view with GADD45A locus displaying ATF4 CUT&RUN and p53 ChIP-Seq data37 in HCT116 WT cell line following 6 h treatment with DMSO, 2 mM histidinol (HisOH), 2 μM tunicamycin (TM), and 5 µM nutlin-3A (NUT). Putative p53RE, ATF4 motif and GADD45A intron 3 enhancer locations are indicated on the bottom, DNaseI hypersensitive sites (DHS) are marked in gray/black. (B) Schematic representation of the GADD45A-nLuc reporter construct with relevant enhancer sequence motifs highlighted in panel C. (D) Normalized luciferase expression values using GADD45A-nLuc reporter transfected into HCT116 WT cell line and 16 h treatment with DMSO, nutlin-3A and tunicamycin as indicated in the legend. Reporter constructs included wild-type, a negative control with 250 bp enhancer deletion (“No Enhancer”) and various ATF4, AP1 and p53RE motif mutations alone or in combination as indicated in the table below (“WT” or “mutant” in gray). Specific mutations in motifs are indicated in panel C. Statistical comparisons were generated using one-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.