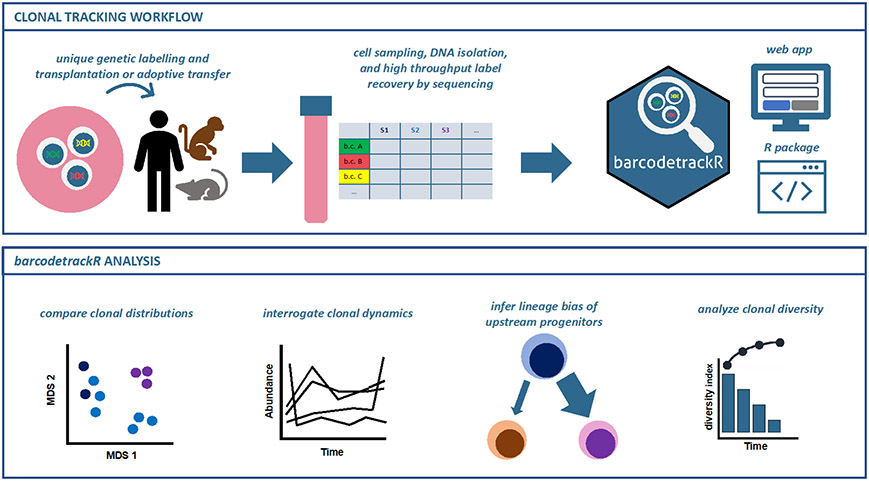
Figure 1: Clonal tracking experimental design and barcodetrackR analysis.
Clonal tracking experiments generally follow the depicted framework, which encompasses, in order, the genetic labelling of cells to create an integrated DNA tag, transplantation or adoptive transfer of these cells into a recipient, acquisition of cellular progeny from the recipient following transplantation or adoptive transfer, genetic tag retrieval from progeny cells followed by high-throughput sequencing, algorithmic quantification of detected individual unique tags, and finally, downstream analyses, where the barcodetrackR toolkit can be utilized (top panel). barcodetrackR contains functions for analyzing relationships between sample clonal distributions, interrogating clonal dynamics over time, inferring pairwise lineage bias of upstream progenitors, and analyzing longitudinal sample clonal diversity (bottom panel).