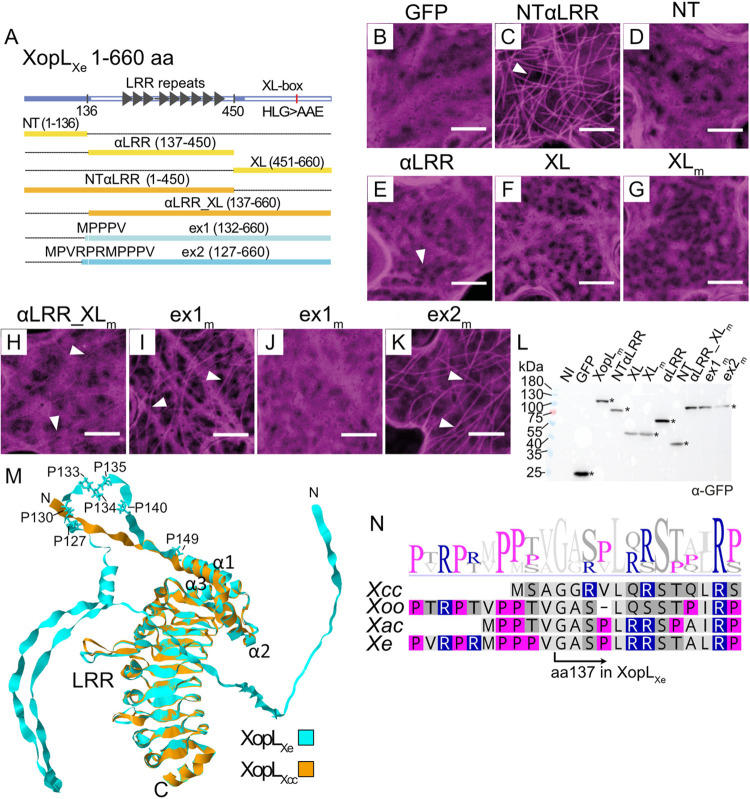
Fig 5. Comparative sequence analysis of XopL homologs identifies a proline-rich-region (PRR) important for MT localization.
(A) Domain structure of XopLXe and derivatives used to functionally analyze the PRR. Domains crystalized by Singer et al. (2013) are denoted with a white bar, LRR repeats by dark gray arrows. Amino acid exchanges in the XL-box of XopLXe to generate XopLm are indicated. Single domain derivatives are annotated in yellow, multiple domain in orange, and ‘extension’ (ex) constructs in blue. Amino acids added to the αLRR_XL in ex1 and ex2 are shown at the N-terminus of the corresponding construct. Amino acid positions included in each derivative are shown in brackets. (B-K) Confocal microscopy pictures of lower epidermal cells of wild-type N. benthamiana leaves. Leaves were agroinfected (OD600 of 0.4) to express (B) a GFP control, (C) NTαLRR, (D) NT, (E) αLRR, (F) XL, (G) XLm, (H) αLRR_XLm, (I) ex1m; cell with MT localization, (J) ex1m; cell without MT localization and (K) ex2m. Subscript ‘m’ indicates a triple amino acid exchange in the E3 ligase domain (H584A, L585A, G586E) that renders the derivative E3 ligase-inactive. Samples were harvested at 2 dpi for microscopy. All XopL derivatives are tagged at the C-terminus with GFP (visible in magenta). Examples of MT association are indicated by white arrows. Scale bars are 5 μm. (L) Western blot analysis of protein extracts isolated 2 dpi from experiment in (B-K). Signals were detected using a GFP-specific antibody (*). The left side of the blot shows protein mass in kDa. (M) An alignment of XopLXcc (orange) and XopLXe (cyan) 3D structural models generated in AlphaFold2. The alignment includes the unstructured NT, α-helical (α1–3) and LRR regions. The cluster of prolines conserved in MT-localizing XopLs are labeled and represented in stick format. N- and C-termini are marked with N and C, respectively. Models do not include the C-terminal XL-box. (N) Alignment of XopLXcc, XopLXe, XopLXac and XopLXoo at the junction between NT and αLRR constructs. Amino acids are colored based on polarity (Geneious Prime), basic amino acids are in blue, and fuchsia highlights prolines. The boundary between the NT and αLRR constructs is annotated at amino acid 137. The sequence logo above the alignment shows sequence conservation at specific positions.