Fig 7. The exchange of the NTα region of XopLXe and XopLXcc is sufficient to engineer a MT-binding XopLXcc.
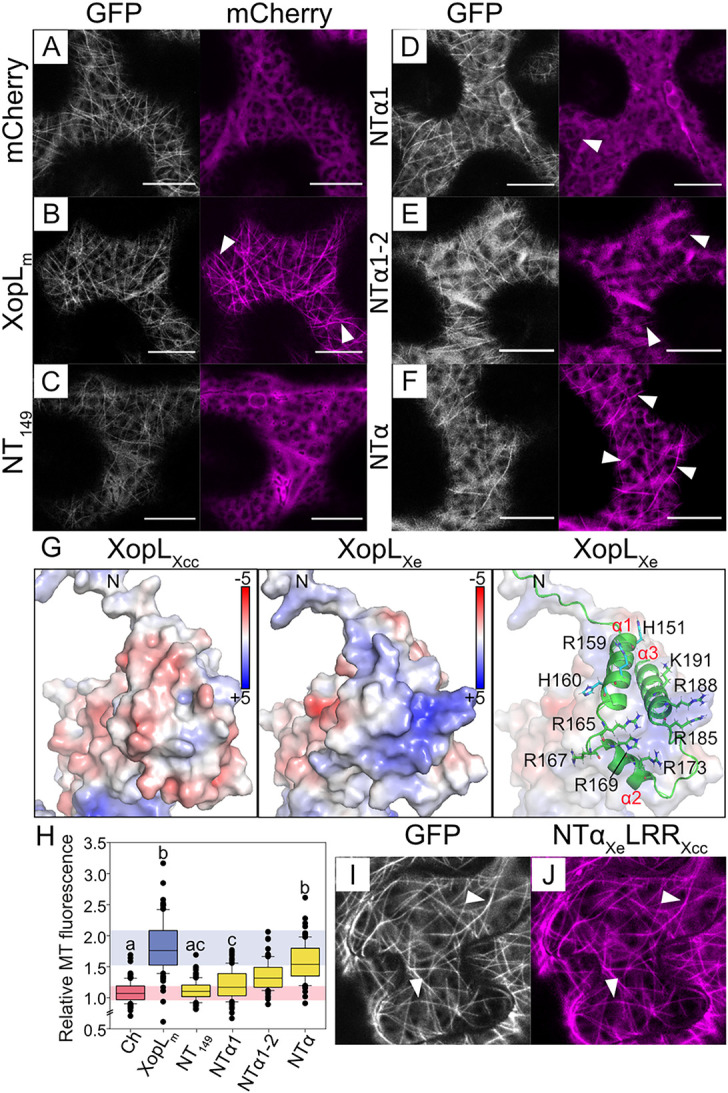
(A-F, H-J) Confocal microscopy images of lower epidermal cells of GFP-TUA6 (labels MTs) transgenic N. benthamiana leaves. Leaves were agroinfected (OD600 of 0.8) to express (A) mCherry and (B) XopLm controls and (C-F) XopL derivatives: (C) XopLXe 1–149 aa (NT149), (D) XopLXe 1–164 aa (NTα1), (E) XopLXe 1–179 aa (NTα1–2), (F) XopLXe 1–210 aa (NTα; α1–3). Scale bars are 15 μm. The mCherry channel is visible in magenta and GFP (MTs) is white. White arrows point out examples of MT labeling. The corresponding western blot is shown in S9 Fig. (G) Electrostatic surface potential comparison of the α-helical regions of XopLXcc and XopLXe modelled with AlphaFold2/PyMOL. Values are expressed on a color scale from -5 (red) to +5 (blue) kT/e. The right panel depicts an overlay of the electrostatic map of XopLXe with its ribbon structure to show the locations of α 1–3. Basic residues are represented in stick format. N-terminal ends of proteins are marked ‘N’ in all panels (H) Quantification of MT-association of XopL derivatives from confocal images of agroinfected GFP-TUA6 lower leaf epidermis (2 dpi, OD600 of 0.8) depicted in (A-F). MT binding is expressed as ‘Relative MT fluorescence’ (see Materials and Methods). Treatments that are not significantly different (p>0.05) are labeled with the same letter (Kruskal-Wallis One Way Analysis of Variance on Ranks, pairwise Tukey post-hoc). Background colors are included for ease of comparison between key controls XopLm (blue) and mCherry (red). Boxes represents first to third quartiles and the median is represented by a horizontal line. Whiskers represent the distribution of remaining data points. (I-J) Confocal microscopy image of XopLXe 1–210 aa translationally fused with XopLXcc 78–428 aa (NTαXeLRRXcc) from the same experiment as depicted in (A-F). (I) GFP labeled MTs are in white, (J) the domain swapped XopL in magenta. White arrows point out examples of MT labeling. Scale bars are 15 μm. The corresponding western blot is shown in S9 Fig.
