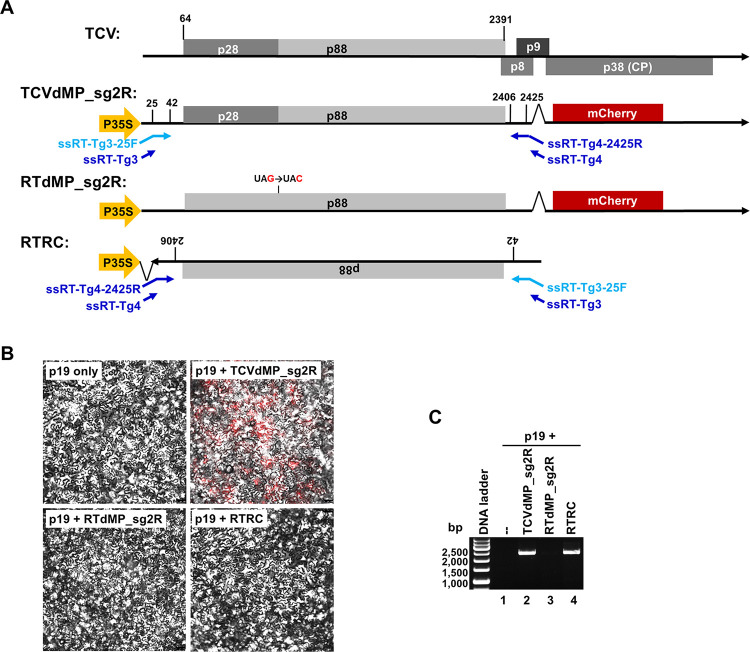
Fig 1. Generation of (-)-strand-specific RT-PCR products of TCV for error profiling.
A. Constructs assembled for the current study. The top diagram depicts the (+)-strand genome of TCV encoding proteins p28, p88, p8, p9, and p38. Among them, p8 and p9 are translated from sgRNA1, p38 sgRNA2. TCVdMP_sg2R is a modified TCV replicon integrated in the binary plasmid pAI101, downstream of the strong P35S promoter so that once inside plant cells, the host cell PolII would be recruited to transcribe replication-initiating viral RNAs. Note that the p8 and p9 MP ORFs were deleted, and the p38 ORF was replaced by that of mCherry. Note that the short, arrowed lines in blue symbolize the four primers used in strand-specific RT-PCR, with the one in light blue used in the RT step, whereas the three in dark blue used in the PCR step. RTdMP_sg2R was further modified from TCVdMP_sg2R by eliminating the p28 stop codon, rendering its transcripts incapable of launching replication, hence are exclusively (+) strands. RTRC contains the reverse-complemented form of the first 2,489 nt of RTdMP_sg2R, so that P35S-driven transcription leads to RNAs that are exclusively (-)-stranded relative to TCV genome. B. Confocal microscopy images of N. benthamiana epidermal cells showing that only TCVdMP_sg2R replicated to produce mCherry fluorescene. C. Strand-specific RT-PCR showing that only the two constructs expected to synthesize (-) strands, TCVdMP_sg2R and RTRC, produced PCR products of expected size (2,363 bp). Note that at 24 PCR cycles, the RTRC-specific product was much less abundant.