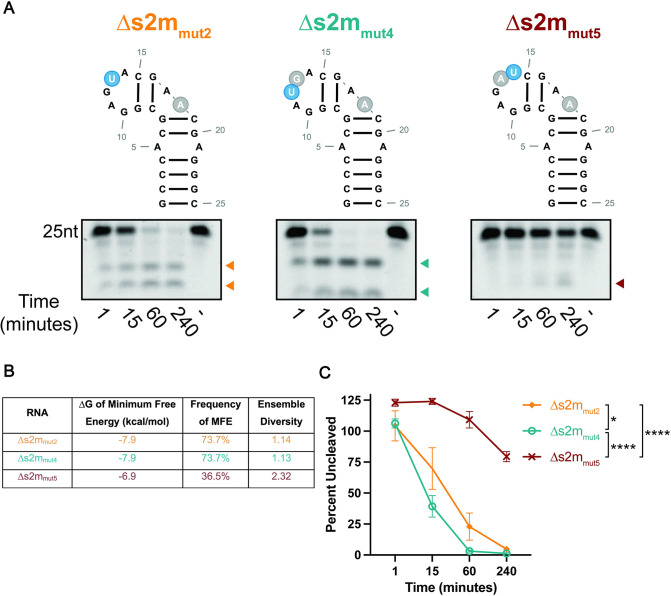
Fig 3. Uracil position in loop structures impacts SARS-CoV-2 nsp15 cleavage efficiency.
A. Endonuclease assays of s2m pentaloop mutant RNAs. Reactions were run at a nsp15 hexamer:RNA ratio of 1:6 run on a 22.5% TBE-Urea Gel. Full-length bands indicated at the top of the gel with cleavage products below. Representative images from one experiment. Cleavage products that accumulate over time are indicated by arrows. B. Thermodynamic stability, frequency of MFE, and ensemble diversity of ∆s2m mutant RNAs indicated as predicted by RNAfold. C. Percent of full-length RNA remaining as measured by densitometry. Percentage of uncut RNA was calculated by normalizing to a denatured nsp15 control (indicated by (-) in the far-right lane). RNAs with uridines positioned closer to the apex of the pentaloop cleaved more rapidly than ∆s2mmut5 in which the uridine is at the end of the pentaloop. AUC was calculated for each RNA, and one-way ANOVA with multiple comparisons was performed on the AUC. Data represents three independent experiments.