FIG 4.
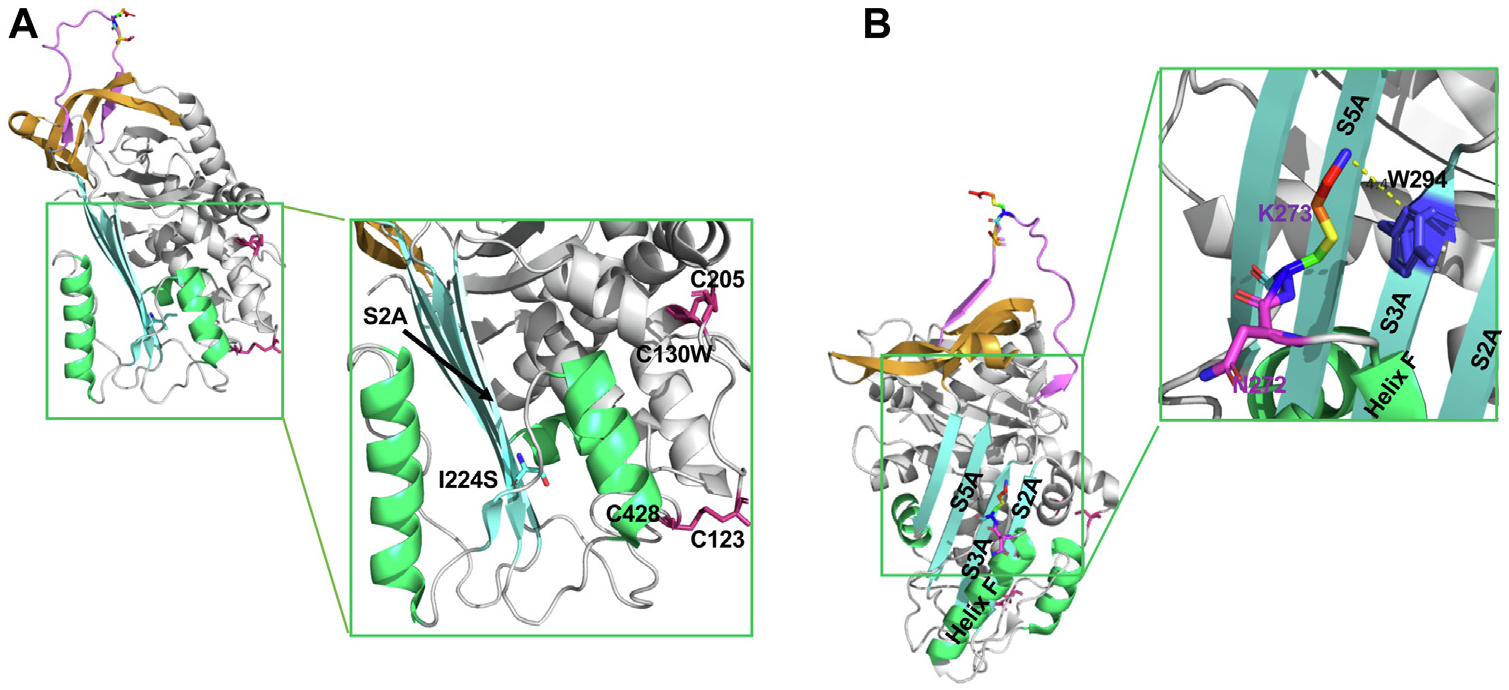
Structural analysis of I224S, N272del, and K273del. A, Structural analysis of I224S. The “reoriented” structure of active C1-INH is shown in a cartoon representation (PDB: 5DU3).7 Isoleucine has a large hydrophobic side chain, which enhances the stability of the hydrophobic core of β-strands in C1-INH. The change of I224 to S destabilizes the hydrophobic core of the antiparallel β-strands (SA) in C1-INH. The 2 disulfide bonds are shown in pink. B, Structural analysis of N272del and K273del. N272 is one of the N-glycosylation sites. The deletion of N272 will likely disrupt the N-linked glycosylation site and impair protein folding. K273 is located in the loop right after helix F. The side chain of K273 forms the stabilizing cation-π interactions with the aromatic side chain from W294. The deletion of K273 will disrupt this interaction and affect the conformation of helix F. K273del previously reported resulting in a new N-glycosylation site in C1-INH.25 The N272 residue is shown in pink and K273 is shown in rainbow. PDB, Protein Data Bank; SA, β-sheets A.
